45a12527d39e33032a5f04f2f52e2f9c.ppt
- Количество слайдов: 33
 A Dynamic Data Driven Grid System for Intra-operative Image Guided Neurosurgery A Majumdar 1, A Birnbaum 1, D Choi 1, A Trivedi 2, S. K. Warfield 3, K. Baldridge 1, and Petr Krysl 2 1 San Diego Supercomputer Center University of California San Diego 2 Structural Engineering Dept University of California San Diego Computational Radiology Lab Brigham and Women’s Hospital Harvard Medical School 3 Grants: NSF: ITR 0427183, 0426558; NIH: P 41 RR 13218, P 01 CA 67165, LM 0078651, I 3 grant (IBM) San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
A Dynamic Data Driven Grid System for Intra-operative Image Guided Neurosurgery A Majumdar 1, A Birnbaum 1, D Choi 1, A Trivedi 2, S. K. Warfield 3, K. Baldridge 1, and Petr Krysl 2 1 San Diego Supercomputer Center University of California San Diego 2 Structural Engineering Dept University of California San Diego Computational Radiology Lab Brigham and Women’s Hospital Harvard Medical School 3 Grants: NSF: ITR 0427183, 0426558; NIH: P 41 RR 13218, P 01 CA 67165, LM 0078651, I 3 grant (IBM) San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 TALK SECTIONS 1. 2. 3. 4. 5. PROBLEM DESCRIPTION AND DDDAS GRID ARCHITECTURE ADVANCED BIOMECHANICAL MODEL PARALLEL AND END-to-END TIMING SUMMARY San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
TALK SECTIONS 1. 2. 3. 4. 5. PROBLEM DESCRIPTION AND DDDAS GRID ARCHITECTURE ADVANCED BIOMECHANICAL MODEL PARALLEL AND END-to-END TIMING SUMMARY San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 1. PROBLEM DESCRIPTION AND DDDAS San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
1. PROBLEM DESCRIPTION AND DDDAS San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 Neurosurgery Challenge • Challenges : • • Remove as much tumor tissue as possible Minimize the removal of healthy tissue Avoid the disruption of critical anatomical structures Know when to stop the resection process • Compounded by the intra-operative brain shape deformation that happens as a result of the surgical process – preoperative plan diminishes • Important to be able to quantify and correct for these deformations while surgery is in progress by dynamically updating pre-operative images in a way that allows surgeons to react to these changing conditions • The simulation pipeline must meet the real-time constraints of neurosurgery – provide images approx. once/hour within few minutes during surgery lasting 6 to 8 hours San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Neurosurgery Challenge • Challenges : • • Remove as much tumor tissue as possible Minimize the removal of healthy tissue Avoid the disruption of critical anatomical structures Know when to stop the resection process • Compounded by the intra-operative brain shape deformation that happens as a result of the surgical process – preoperative plan diminishes • Important to be able to quantify and correct for these deformations while surgery is in progress by dynamically updating pre-operative images in a way that allows surgeons to react to these changing conditions • The simulation pipeline must meet the real-time constraints of neurosurgery – provide images approx. once/hour within few minutes during surgery lasting 6 to 8 hours San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
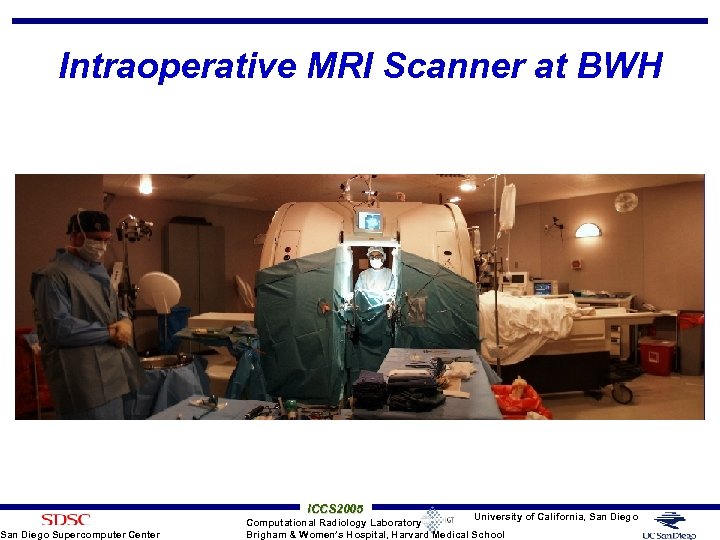 Intraoperative MRI Scanner at BWH San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Intraoperative MRI Scanner at BWH San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
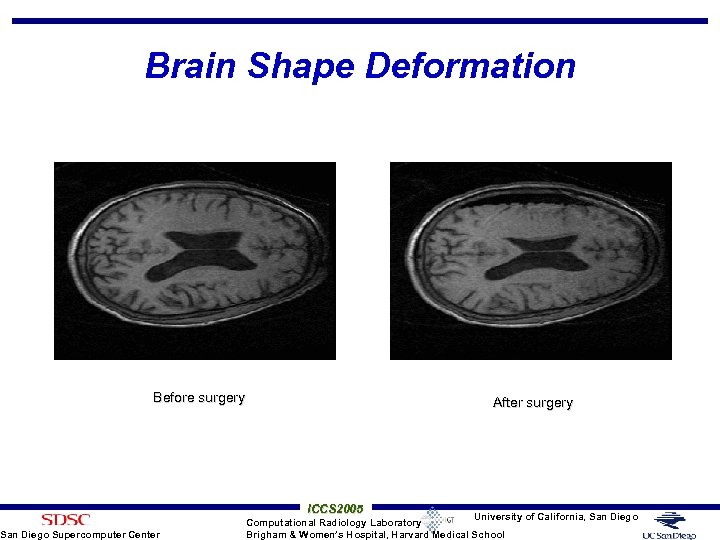 Brain Shape Deformation Before surgery San Diego Supercomputer Center After surgery ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Brain Shape Deformation Before surgery San Diego Supercomputer Center After surgery ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
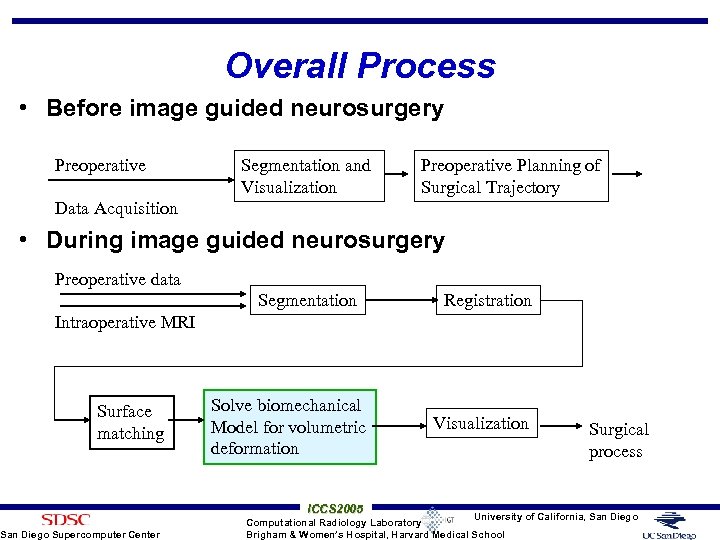 Overall Process • Before image guided neurosurgery Preoperative Segmentation and Visualization Preoperative Planning of Surgical Trajectory Data Acquisition • During image guided neurosurgery Preoperative data Segmentation Registration Intraoperative MRI Surface matching San Diego Supercomputer Center Solve biomechanical Model for volumetric deformation ICCS 2005 Visualization Surgical process University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Overall Process • Before image guided neurosurgery Preoperative Segmentation and Visualization Preoperative Planning of Surgical Trajectory Data Acquisition • During image guided neurosurgery Preoperative data Segmentation Registration Intraoperative MRI Surface matching San Diego Supercomputer Center Solve biomechanical Model for volumetric deformation ICCS 2005 Visualization Surgical process University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
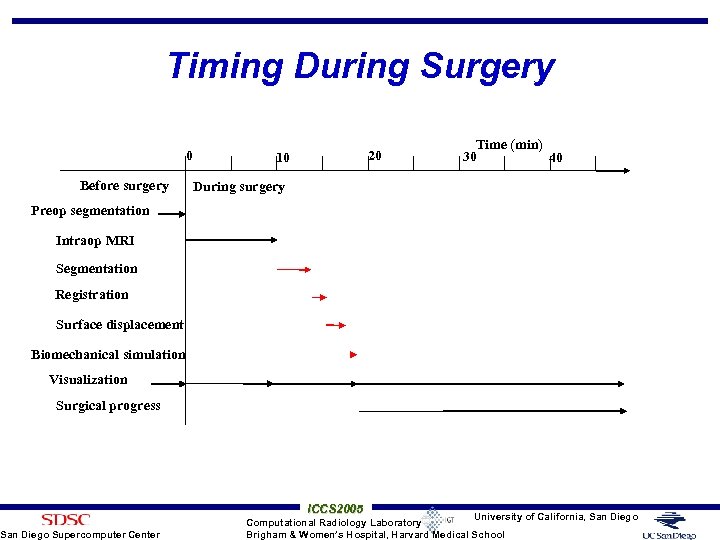 Timing During Surgery 0 Before surgery 20 10 Time (min) 30 40 During surgery Preop segmentation Intraop MRI Segmentation Registration Surface displacement Biomechanical simulation Visualization Surgical progress San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Timing During Surgery 0 Before surgery 20 10 Time (min) 30 40 During surgery Preop segmentation Intraop MRI Segmentation Registration Surface displacement Biomechanical simulation Visualization Surgical progress San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
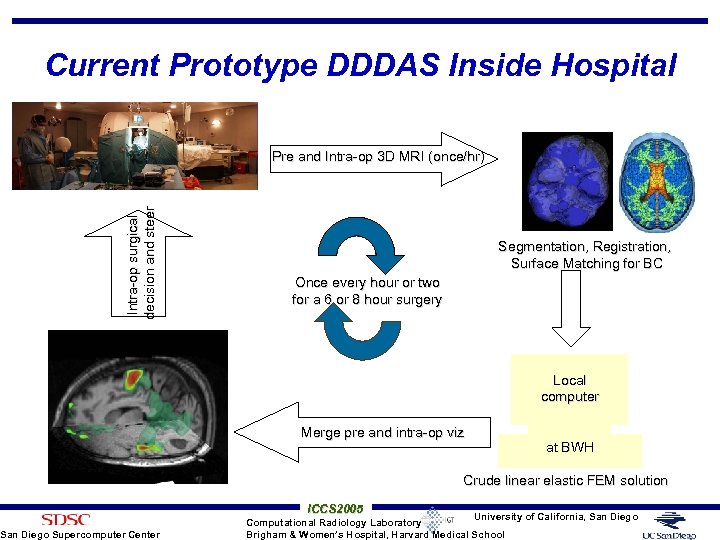 Current Prototype DDDAS Inside Hospital Intra-op surgical decision and steer Pre and Intra-op 3 D MRI (once/hr) San Diego Supercomputer Center Segmentation, Registration, Surface Matching for BC Once every hour or two for a 6 or 8 hour surgery Local computer Merge pre and intra-op viz at BWH Crude linear elastic FEM solution ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Current Prototype DDDAS Inside Hospital Intra-op surgical decision and steer Pre and Intra-op 3 D MRI (once/hr) San Diego Supercomputer Center Segmentation, Registration, Surface Matching for BC Once every hour or two for a 6 or 8 hour surgery Local computer Merge pre and intra-op viz at BWH Crude linear elastic FEM solution ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 Current Prototype DDDAS System • Receives 3 -D MRI from operating room once/hour or so • Uses displacement of known surface points as BC to solve a crude linear elastic biomechanical FEM material model on compute system located at BWH • This crude inaccurate model is solvable within the time constraint of few minutes once an hour on local computers at BWH • Dynamically updates pre-op images with biomechanical volumetric simulation based intra-op images • Time critical updates shown to surgeons for intra-op surgical navigation San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Current Prototype DDDAS System • Receives 3 -D MRI from operating room once/hour or so • Uses displacement of known surface points as BC to solve a crude linear elastic biomechanical FEM material model on compute system located at BWH • This crude inaccurate model is solvable within the time constraint of few minutes once an hour on local computers at BWH • Dynamically updates pre-op images with biomechanical volumetric simulation based intra-op images • Time critical updates shown to surgeons for intra-op surgical navigation San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 Two Research Aspects • Grid Architecture – grid scheduling, on demand remote access to multi-teraflop machines, data transfer • Data transfer from BWH to SDSC, solution of detail advanced biomechanical model, transfer of results back to BWH for visualization need to be performed in a few minutes • Development of detailed advanced non-linear scalable viscoelastic biomechanical model • To capture detail intraoperative brain deformation San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Two Research Aspects • Grid Architecture – grid scheduling, on demand remote access to multi-teraflop machines, data transfer • Data transfer from BWH to SDSC, solution of detail advanced biomechanical model, transfer of results back to BWH for visualization need to be performed in a few minutes • Development of detailed advanced non-linear scalable viscoelastic biomechanical model • To capture detail intraoperative brain deformation San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
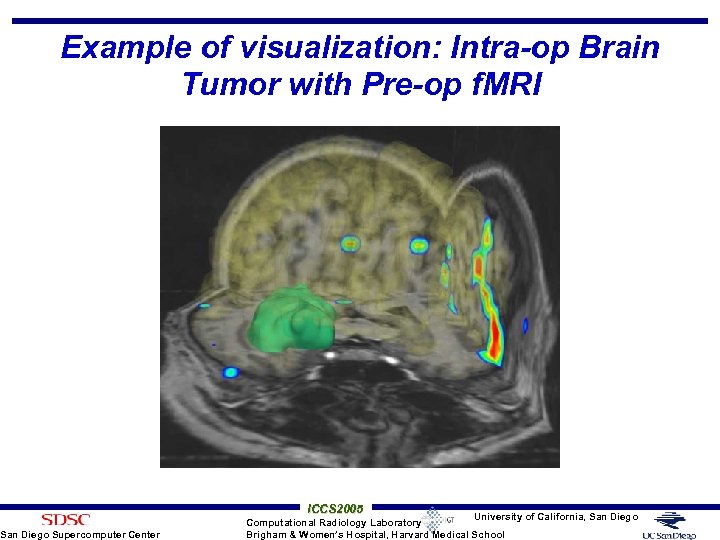 Example of visualization: Intra-op Brain Tumor with Pre-op f. MRI San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Example of visualization: Intra-op Brain Tumor with Pre-op f. MRI San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 2. GRID ARCHITECTURE San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
2. GRID ARCHITECTURE San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 Queue Delay Experiment on Tera. Grid Cluster • Tera. Grid is a NSF funded grid infrastructure across multiple research and academic sites • Queue delays at SDSC and NCSA TG were measured over 3 days for 5 mins wall clock time on 2 to 64 CPUs • Single job submitted at a time • If job didn’t start within 10 mins, job terminated, next one processed • What is the likelihood of job running • 313 jobs to NCSA TG cluster and 332 to SDSC TG cluster – 50 to 56 jobs of each size on each cluster San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Queue Delay Experiment on Tera. Grid Cluster • Tera. Grid is a NSF funded grid infrastructure across multiple research and academic sites • Queue delays at SDSC and NCSA TG were measured over 3 days for 5 mins wall clock time on 2 to 64 CPUs • Single job submitted at a time • If job didn’t start within 10 mins, job terminated, next one processed • What is the likelihood of job running • 313 jobs to NCSA TG cluster and 332 to SDSC TG cluster – 50 to 56 jobs of each size on each cluster San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
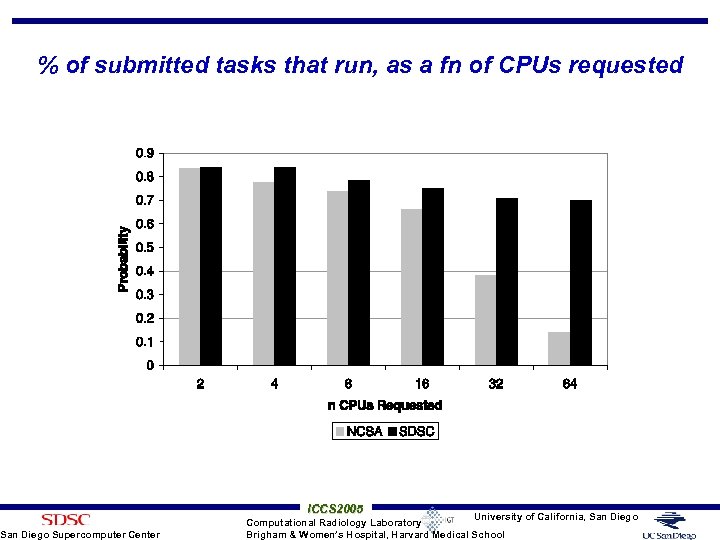 % of submitted tasks that run, as a fn of CPUs requested San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
% of submitted tasks that run, as a fn of CPUs requested San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
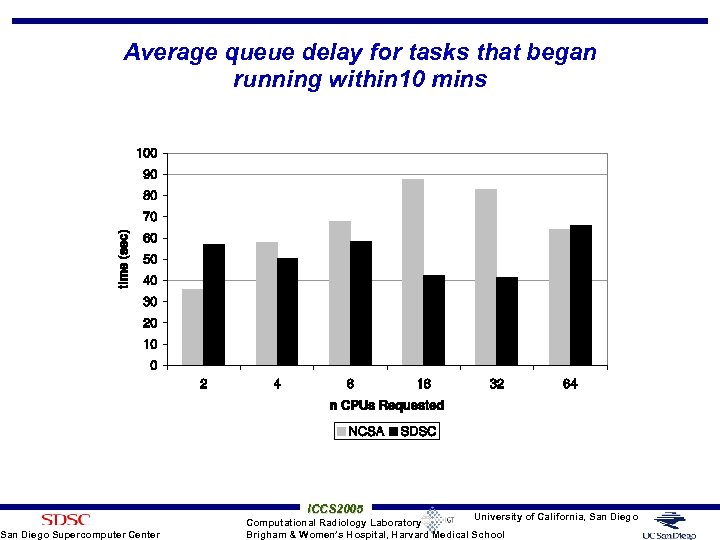 Average queue delay for tasks that began running within 10 mins San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Average queue delay for tasks that began running within 10 mins San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 Queue Delay Test Conclusion • There appears to be a direct relationship between the size of request and the length of the queue delay • Two clusters exhibit different performance profiles • This behavior of queue systems clearly merits further study • More rigorous statistical characterization ongoig on much larger data sets San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Queue Delay Test Conclusion • There appears to be a direct relationship between the size of request and the length of the queue delay • Two clusters exhibit different performance profiles • This behavior of queue systems clearly merits further study • More rigorous statistical characterization ongoig on much larger data sets San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
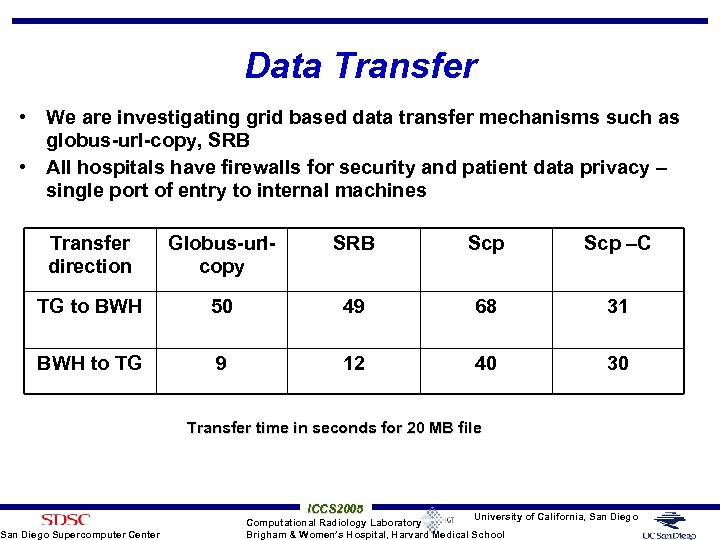 Data Transfer • We are investigating grid based data transfer mechanisms such as globus-url-copy, SRB • All hospitals have firewalls for security and patient data privacy – single port of entry to internal machines Transfer direction Globus-urlcopy SRB Scp –C TG to BWH 50 49 68 31 BWH to TG 9 12 40 30 San Diego Supercomputer Center Transfer time in seconds for 20 MB file ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Data Transfer • We are investigating grid based data transfer mechanisms such as globus-url-copy, SRB • All hospitals have firewalls for security and patient data privacy – single port of entry to internal machines Transfer direction Globus-urlcopy SRB Scp –C TG to BWH 50 49 68 31 BWH to TG 9 12 40 30 San Diego Supercomputer Center Transfer time in seconds for 20 MB file ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 3. ADVANCED BIOMECHANICAL MODEL San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
3. ADVANCED BIOMECHANICAL MODEL San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
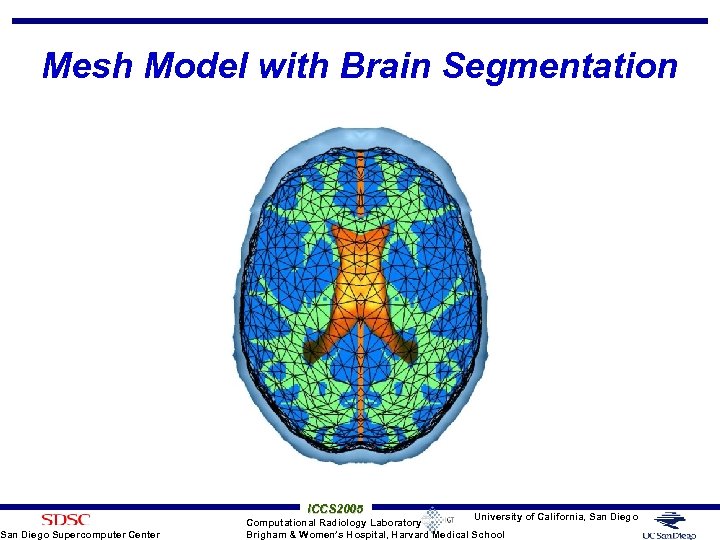 Mesh Model with Brain Segmentation San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Mesh Model with Brain Segmentation San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 Current and New Biomechanical Model • Current linear elastic material model – RTBM • Advanced model under development - FAMULS • Advanced model is based on conforming adaptive refinement method – FAMULS package (AMR) • Inspired by theory of wavelets this refinement produces globally compatible meshes by construction • First task is to replicate the linear elastic result produced by the RTBM code using FAMULS San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Current and New Biomechanical Model • Current linear elastic material model – RTBM • Advanced model under development - FAMULS • Advanced model is based on conforming adaptive refinement method – FAMULS package (AMR) • Inspired by theory of wavelets this refinement produces globally compatible meshes by construction • First task is to replicate the linear elastic result produced by the RTBM code using FAMULS San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
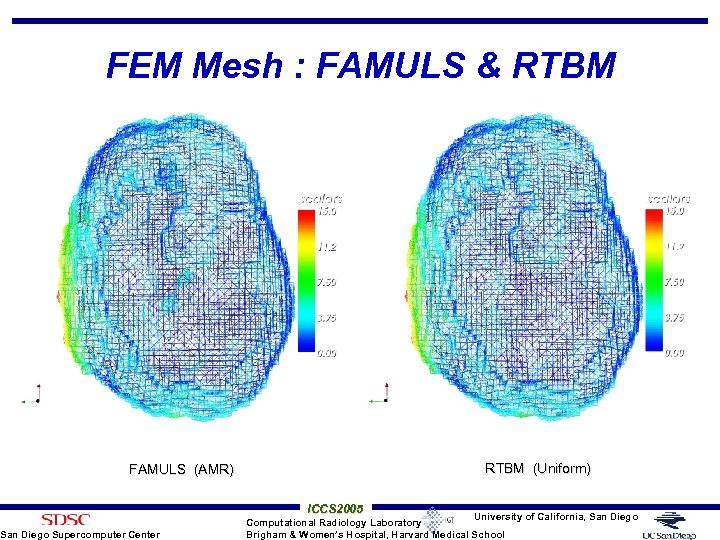 FEM Mesh : FAMULS & RTBM (Uniform) FAMULS (AMR) San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
FEM Mesh : FAMULS & RTBM (Uniform) FAMULS (AMR) San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
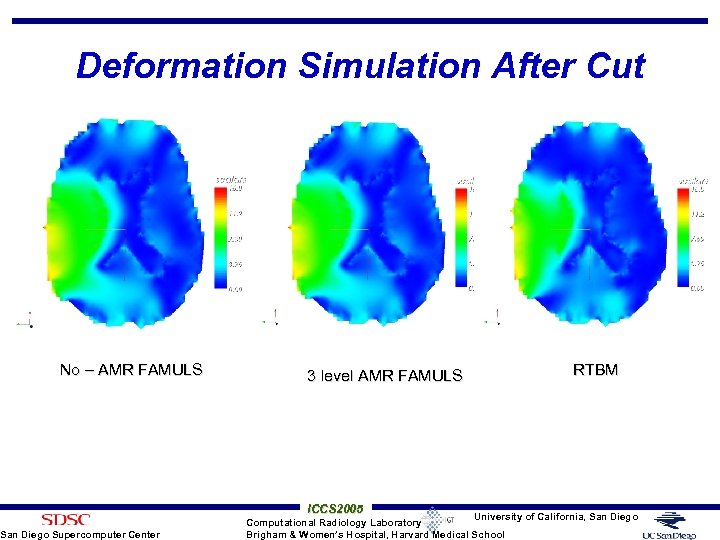 Deformation Simulation After Cut No – AMR FAMULS San Diego Supercomputer Center 3 level AMR FAMULS ICCS 2005 RTBM University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Deformation Simulation After Cut No – AMR FAMULS San Diego Supercomputer Center 3 level AMR FAMULS ICCS 2005 RTBM University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 Advanced Biomechanical Model • The current solver is based on small strain isotropic elastic principle • The new biomechanical model will be inhomogeneous scalable non-linear viscoelastic model with AMR • We also want to increase resolution close to the level of MRI voxels i. e. millions of FEM meshes • Since this complex model still has to meet the real time constraint of neurosurgery it requires fast access to remote multi-tflop systems San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Advanced Biomechanical Model • The current solver is based on small strain isotropic elastic principle • The new biomechanical model will be inhomogeneous scalable non-linear viscoelastic model with AMR • We also want to increase resolution close to the level of MRI voxels i. e. millions of FEM meshes • Since this complex model still has to meet the real time constraint of neurosurgery it requires fast access to remote multi-tflop systems San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 4. PARALLEL AND END-to-END TIMING San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
4. PARALLEL AND END-to-END TIMING San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
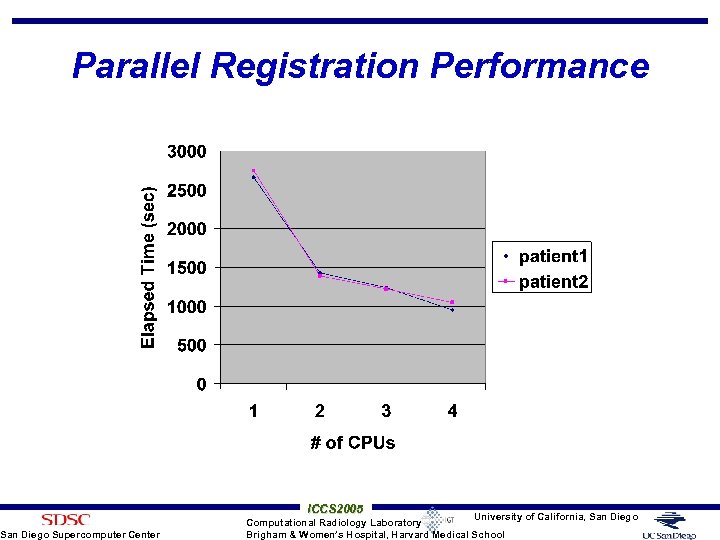 Parallel Registration Performance San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Parallel Registration Performance San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
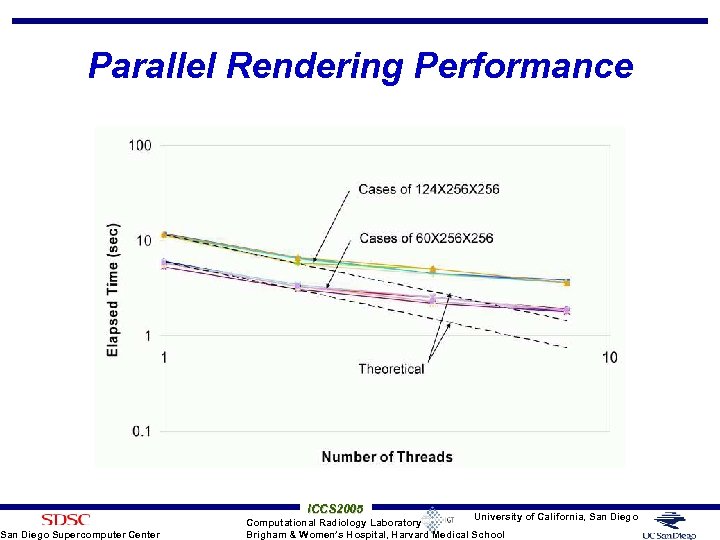 Parallel Rendering Performance San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Parallel Rendering Performance San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
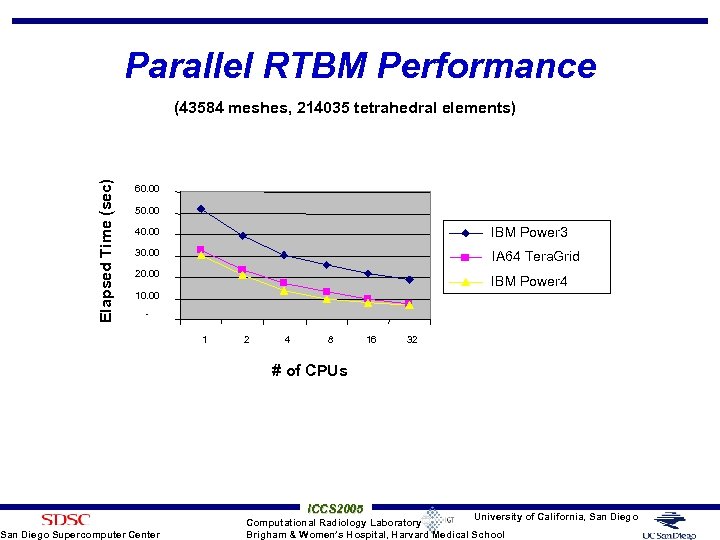 Parallel RTBM Performance Elapsed Time (sec) (43584 meshes, 214035 tetrahedral elements) 60. 00 50. 00 40. 00 IBM Power 3 30. 00 IA 64 Tera. Grid 20. 00 IBM Power 4 10. 00 - San Diego Supercomputer Center 1 2 4 8 16 32 # of CPUs ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Parallel RTBM Performance Elapsed Time (sec) (43584 meshes, 214035 tetrahedral elements) 60. 00 50. 00 40. 00 IBM Power 3 30. 00 IA 64 Tera. Grid 20. 00 IBM Power 4 10. 00 - San Diego Supercomputer Center 1 2 4 8 16 32 # of CPUs ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 End to End (BWH SDSC BWH) Timing • RTBM – not during surgery • Rendering - during Surgery San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
End to End (BWH SDSC BWH) Timing • RTBM – not during surgery • Rendering - during Surgery San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 End-to-end Timing of RTBM • Timing of transferring ~20 MB files from BWH to SDSC, running simulations on 16 nodes (32 procs), transferring files back to BWH = 9* + (60** + 7***) + 50* = 124 sec. • This shows that the grid infrastructure can provide biomechanical brain deformation simulation solutions (using the linear elastic model) to surgery rooms at BWH within ~ 2 mins using TG machines • This satisfies the tight time constraint set by the neurosurgeons San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
End-to-end Timing of RTBM • Timing of transferring ~20 MB files from BWH to SDSC, running simulations on 16 nodes (32 procs), transferring files back to BWH = 9* + (60** + 7***) + 50* = 124 sec. • This shows that the grid infrastructure can provide biomechanical brain deformation simulation solutions (using the linear elastic model) to surgery rooms at BWH within ~ 2 mins using TG machines • This satisfies the tight time constraint set by the neurosurgeons San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 End-to-end Timing of Rendering DURING SURGERY • MRI data from BWH was transferred to SDSC during a surgery • Parallel rendering was performed at SDSC • Rendered viz was sent back to BWH (but not shown to surgeons) • Total time (for two sets of data) in sec = 2*53 (BWH to SDSC) + 2* 7. 4 (render on 32 procs) + 0. 2 (overlapping viz) + 13. 7 (SDSC to BWH) = 148. 4 sec San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
End-to-end Timing of Rendering DURING SURGERY • MRI data from BWH was transferred to SDSC during a surgery • Parallel rendering was performed at SDSC • Rendered viz was sent back to BWH (but not shown to surgeons) • Total time (for two sets of data) in sec = 2*53 (BWH to SDSC) + 2* 7. 4 (render on 32 procs) + 0. 2 (overlapping viz) + 13. 7 (SDSC to BWH) = 148. 4 sec San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 San Diego Supercomputer Center 5. SUMMARY ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
San Diego Supercomputer Center 5. SUMMARY ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
 Ongoing and Future DDDAS Research • Continuing research and development in grid architecture, on demand computing, data transfer • Continuing development of advanced biomechanical model and parallel algorithm • Moving towards near-continuous DDDAS instead of once an hour or so 3 -D MRI based DDDAS • Scanner at BWH can provide one 2 -D slice every 3 sec or three orthogonal 2 -D slices every 6 sec • Near-continuous DDDAS architecture • Requires major research, development and implementation work in the biomechanical application domain • Requires research in the closed loop system of dynamic image driven continuous biomechanical simulation and 3 -D volumetric FEM results based surgical navigation and steering San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School
Ongoing and Future DDDAS Research • Continuing research and development in grid architecture, on demand computing, data transfer • Continuing development of advanced biomechanical model and parallel algorithm • Moving towards near-continuous DDDAS instead of once an hour or so 3 -D MRI based DDDAS • Scanner at BWH can provide one 2 -D slice every 3 sec or three orthogonal 2 -D slices every 6 sec • Near-continuous DDDAS architecture • Requires major research, development and implementation work in the biomechanical application domain • Requires research in the closed loop system of dynamic image driven continuous biomechanical simulation and 3 -D volumetric FEM results based surgical navigation and steering San Diego Supercomputer Center ICCS 2005 University of California, San Diego Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School


