0b777cd34debb8e03271ea1c0c7a0b52.ppt
- Количество слайдов: 22
 A coalescent computational platform to predict strength of association for clinical samples Genomic studies and the Hap. Map March 15 -18, 2005 Oxford, United Kingdom Gabor T. Marth Department of Biology, Boston College marth@bc. edu
A coalescent computational platform to predict strength of association for clinical samples Genomic studies and the Hap. Map March 15 -18, 2005 Oxford, United Kingdom Gabor T. Marth Department of Biology, Boston College marth@bc. edu
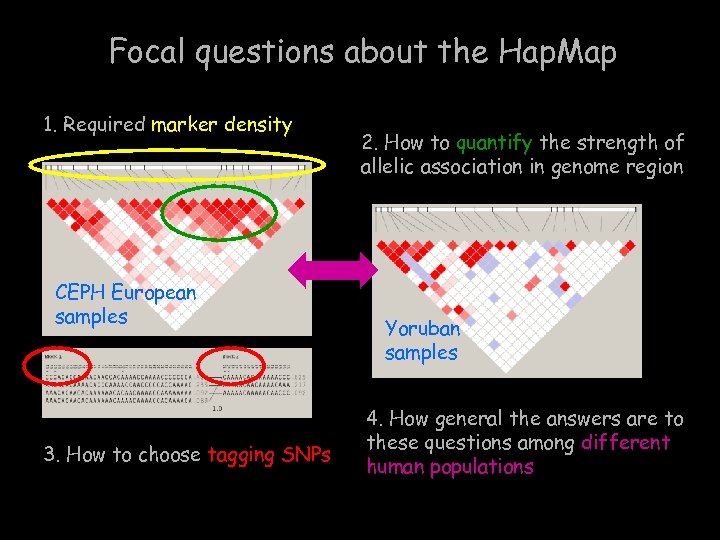 Focal questions about the Hap. Map 1. Required marker density CEPH European samples 3. How to choose tagging SNPs 2. How to quantify the strength of allelic association in genome region Yoruban samples 4. How general the answers are to these questions among different human populations
Focal questions about the Hap. Map 1. Required marker density CEPH European samples 3. How to choose tagging SNPs 2. How to quantify the strength of allelic association in genome region Yoruban samples 4. How general the answers are to these questions among different human populations
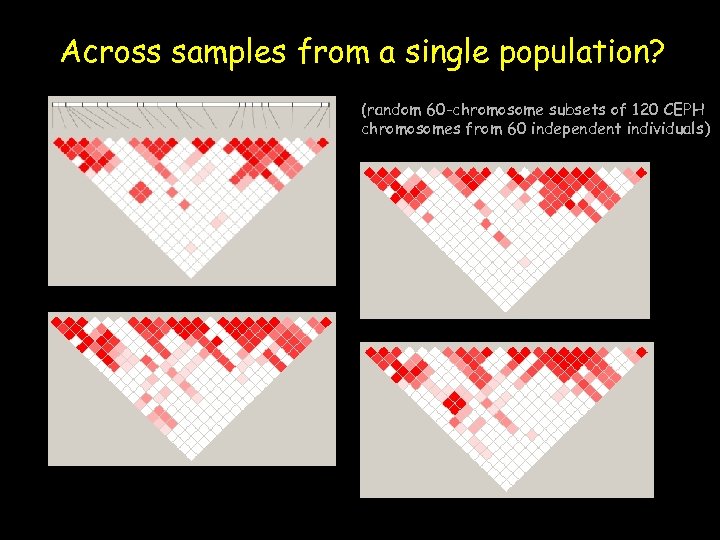 Across samples from a single population? (random 60 -chromosome subsets of 120 CEPH chromosomes from 60 independent individuals)
Across samples from a single population? (random 60 -chromosome subsets of 120 CEPH chromosomes from 60 independent individuals)
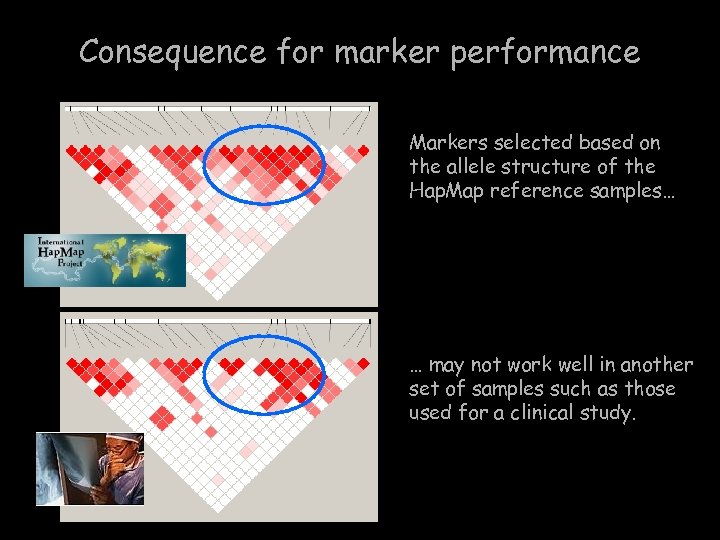 Consequence for marker performance Markers selected based on the allele structure of the Hap. Map reference samples… … may not work well in another set of samples such as those used for a clinical study.
Consequence for marker performance Markers selected based on the allele structure of the Hap. Map reference samples… … may not work well in another set of samples such as those used for a clinical study.
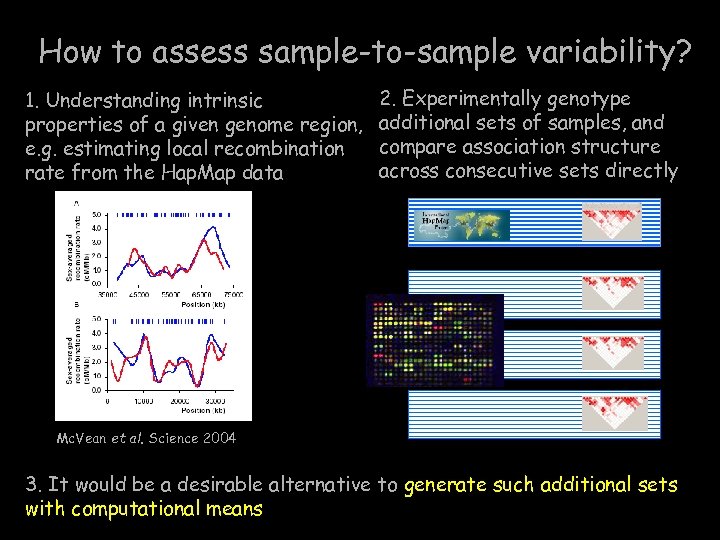 How to assess sample-to-sample variability? 1. Understanding intrinsic properties of a given genome region, e. g. estimating local recombination rate from the Hap. Map data 2. Experimentally genotype additional sets of samples, and compare association structure across consecutive sets directly Mc. Vean et al. Science 2004 3. It would be a desirable alternative to generate such additional sets with computational means
How to assess sample-to-sample variability? 1. Understanding intrinsic properties of a given genome region, e. g. estimating local recombination rate from the Hap. Map data 2. Experimentally genotype additional sets of samples, and compare association structure across consecutive sets directly Mc. Vean et al. Science 2004 3. It would be a desirable alternative to generate such additional sets with computational means
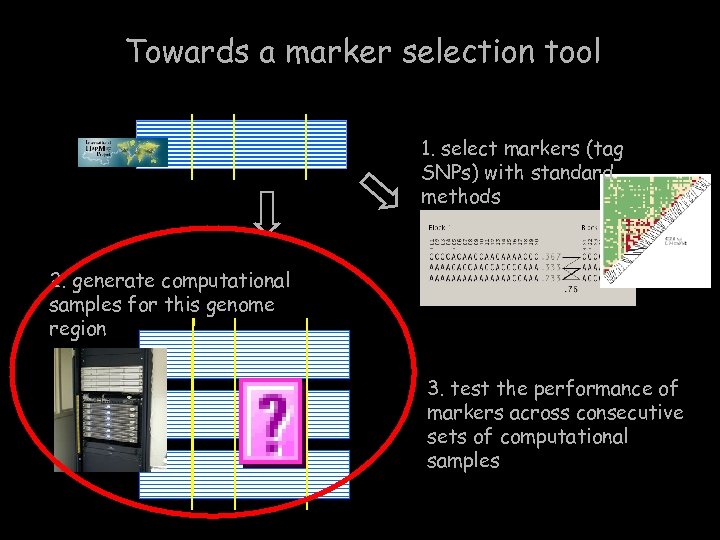 Towards a marker selection tool 1. select markers (tag SNPs) with standard methods 2. generate computational samples for this genome region 3. test the performance of markers across consecutive sets of computational samples
Towards a marker selection tool 1. select markers (tag SNPs) with standard methods 2. generate computational samples for this genome region 3. test the performance of markers across consecutive sets of computational samples
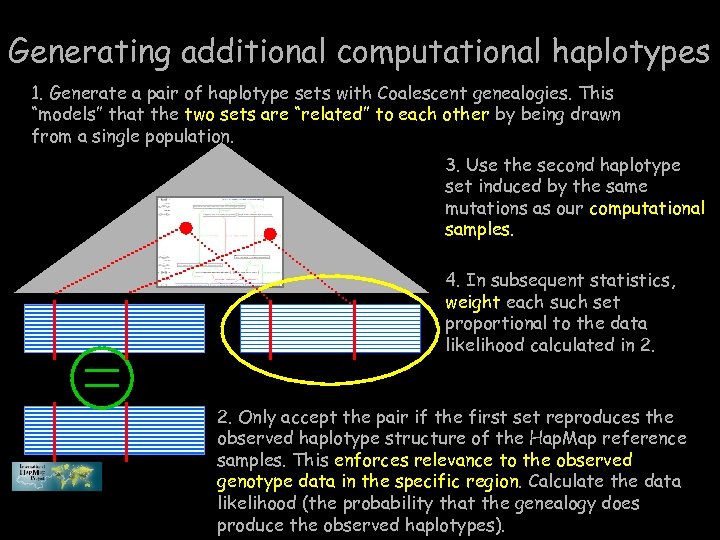 Generating additional computational haplotypes 1. Generate a pair of haplotype sets with Coalescent genealogies. This “models” that the two sets are “related” to each other by being drawn from a single population. 3. Use the second haplotype set induced by the same mutations as our computational samples. 4. In subsequent statistics, weight each such set proportional to the data likelihood calculated in 2. Only accept the pair if the first set reproduces the observed haplotype structure of the Hap. Map reference samples. This enforces relevance to the observed genotype data in the specific region. Calculate the data likelihood (the probability that the genealogy does produce the observed haplotypes).
Generating additional computational haplotypes 1. Generate a pair of haplotype sets with Coalescent genealogies. This “models” that the two sets are “related” to each other by being drawn from a single population. 3. Use the second haplotype set induced by the same mutations as our computational samples. 4. In subsequent statistics, weight each such set proportional to the data likelihood calculated in 2. Only accept the pair if the first set reproduces the observed haplotype structure of the Hap. Map reference samples. This enforces relevance to the observed genotype data in the specific region. Calculate the data likelihood (the probability that the genealogy does produce the observed haplotypes).
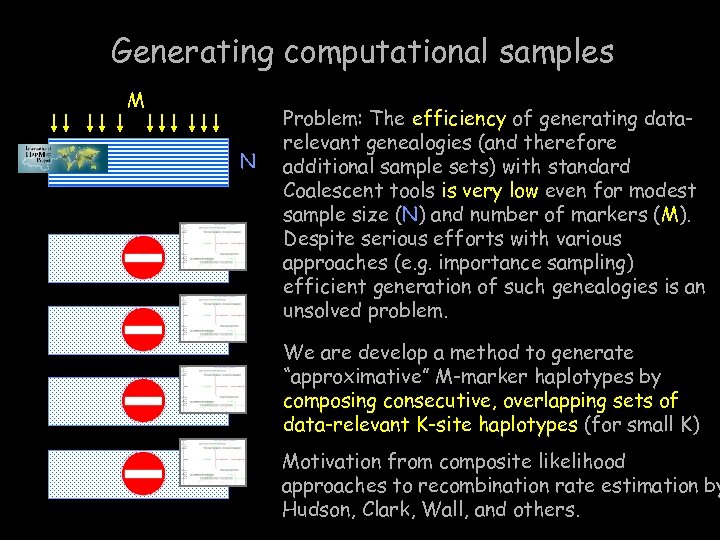 Generating computational samples M N Problem: The efficiency of generating datarelevant genealogies (and therefore additional sample sets) with standard Coalescent tools is very low even for modest sample size (N) and number of markers (M). Despite serious efforts with various approaches (e. g. importance sampling) efficient generation of such genealogies is an unsolved problem. We are develop a method to generate “approximative” M-marker haplotypes by composing consecutive, overlapping sets of data-relevant K-site haplotypes (for small K) Motivation from composite likelihood approaches to recombination rate estimation by Hudson, Clark, Wall, and others.
Generating computational samples M N Problem: The efficiency of generating datarelevant genealogies (and therefore additional sample sets) with standard Coalescent tools is very low even for modest sample size (N) and number of markers (M). Despite serious efforts with various approaches (e. g. importance sampling) efficient generation of such genealogies is an unsolved problem. We are develop a method to generate “approximative” M-marker haplotypes by composing consecutive, overlapping sets of data-relevant K-site haplotypes (for small K) Motivation from composite likelihood approaches to recombination rate estimation by Hudson, Clark, Wall, and others.
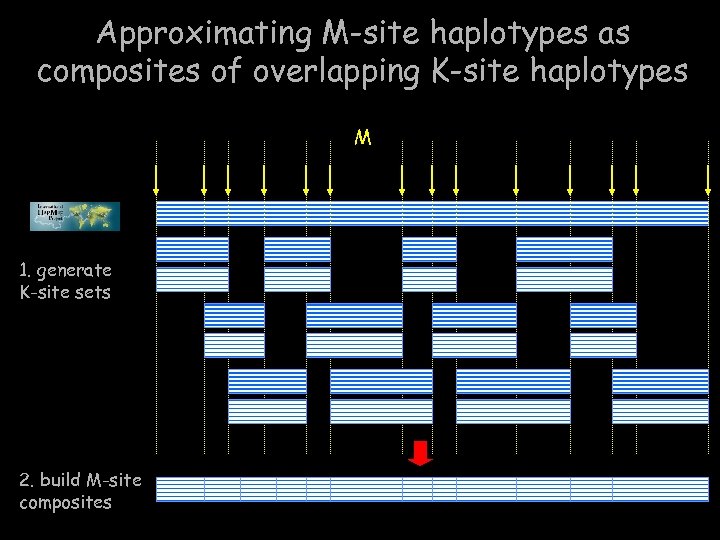 Approximating M-site haplotypes as composites of overlapping K-site haplotypes M 1. generate K-site sets 2. build M-site composites
Approximating M-site haplotypes as composites of overlapping K-site haplotypes M 1. generate K-site sets 2. build M-site composites
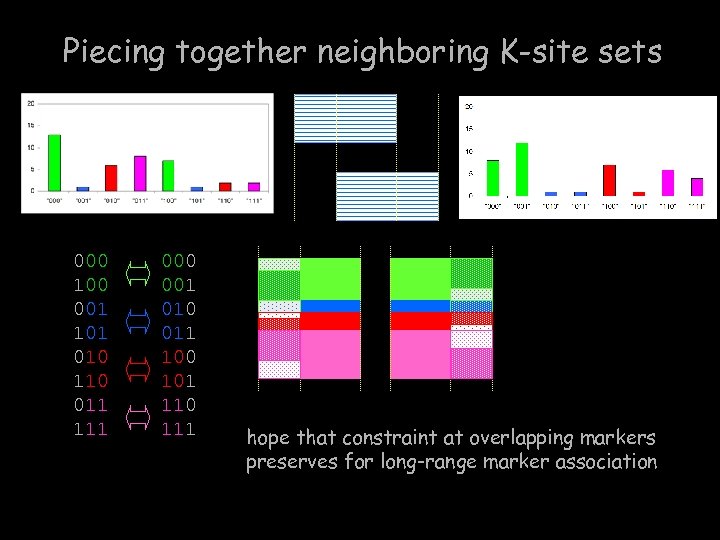 Piecing together neighboring K-site sets 000 100 001 101 010 110 011 111 000 001 010 011 100 101 110 111 hope that constraint at overlapping markers preserves for long-range marker association
Piecing together neighboring K-site sets 000 100 001 101 010 110 011 111 000 001 010 011 100 101 110 111 hope that constraint at overlapping markers preserves for long-range marker association
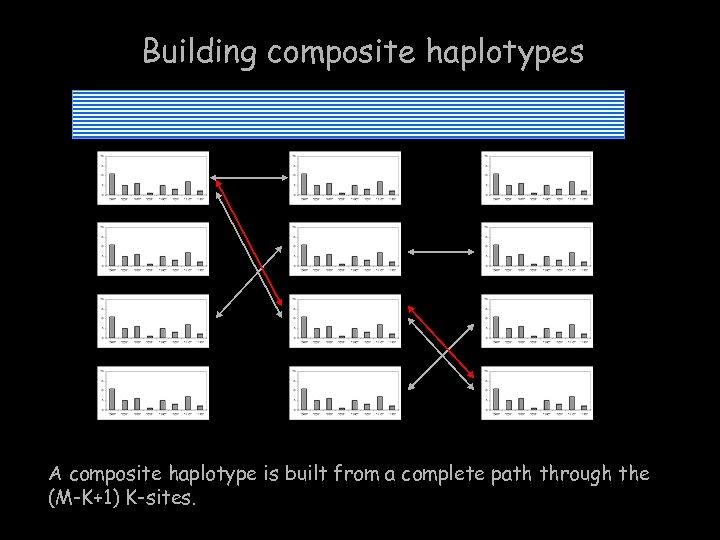 Building composite haplotypes A composite haplotype is built from a complete path through the (M-K+1) K-sites.
Building composite haplotypes A composite haplotype is built from a complete path through the (M-K+1) K-sites.
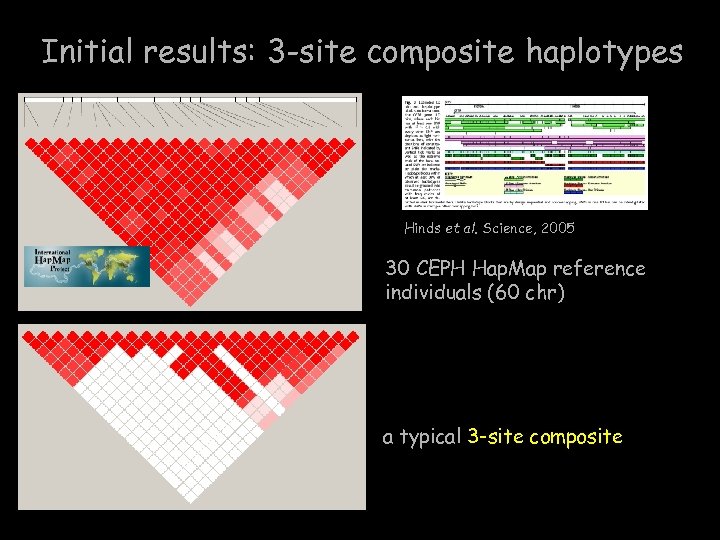 Initial results: 3 -site composite haplotypes Hinds et al. Science, 2005 30 CEPH Hap. Map reference individuals (60 chr) a typical 3 -site composite
Initial results: 3 -site composite haplotypes Hinds et al. Science, 2005 30 CEPH Hap. Map reference individuals (60 chr) a typical 3 -site composite
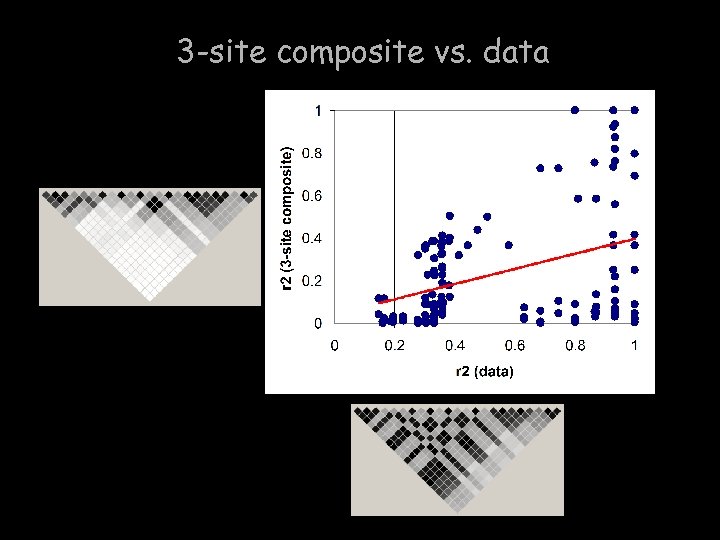 3 -site composite vs. data
3 -site composite vs. data
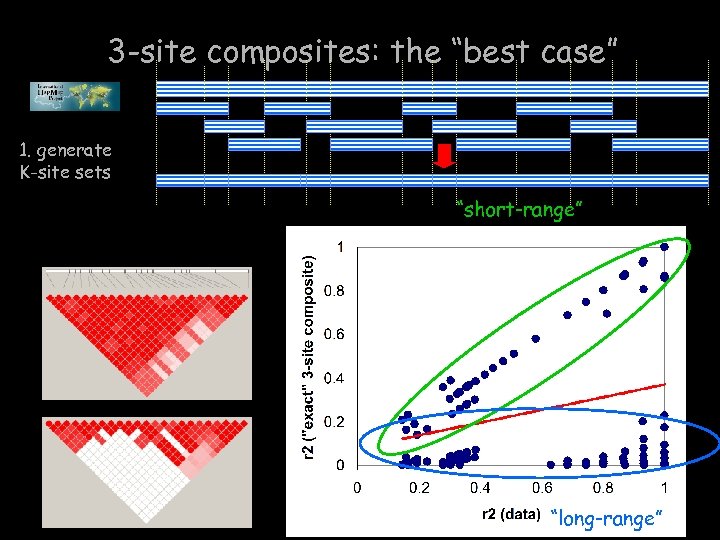 3 -site composites: the “best case” 1. generate K-site sets “short-range” “long-range”
3 -site composites: the “best case” 1. generate K-site sets “short-range” “long-range”
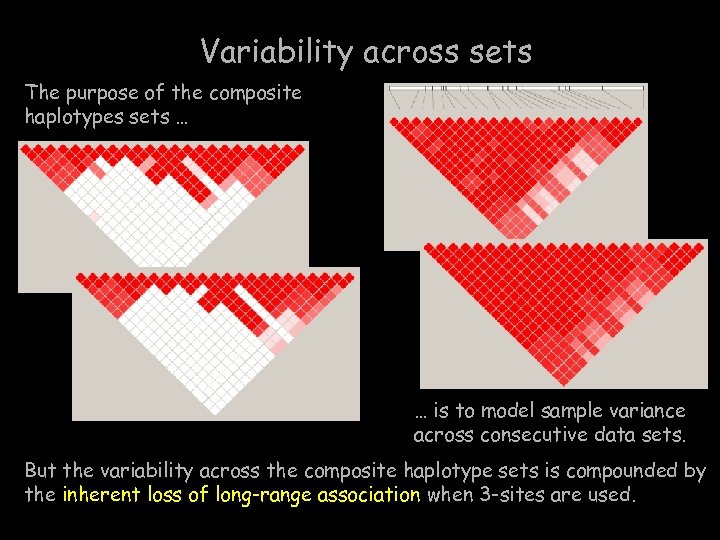 Variability across sets The purpose of the composite haplotypes sets … … is to model sample variance across consecutive data sets. But the variability across the composite haplotype sets is compounded by the inherent loss of long-range association when 3 -sites are used.
Variability across sets The purpose of the composite haplotypes sets … … is to model sample variance across consecutive data sets. But the variability across the composite haplotype sets is compounded by the inherent loss of long-range association when 3 -sites are used.
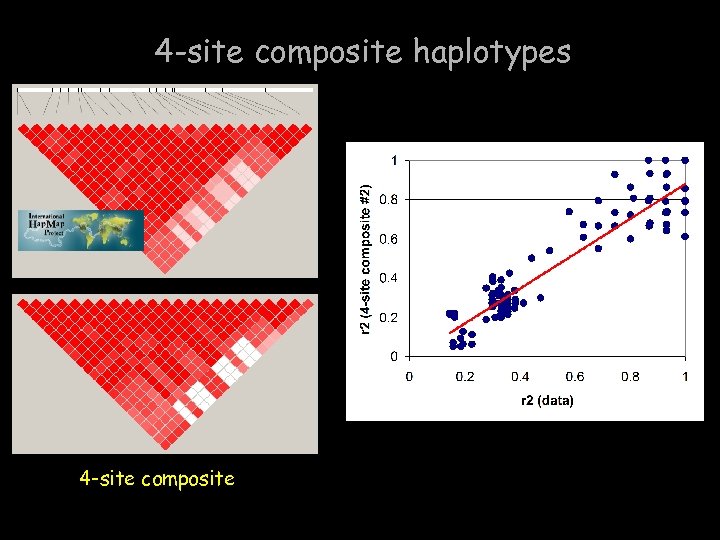 4 -site composite haplotypes 4 -site composite
4 -site composite haplotypes 4 -site composite
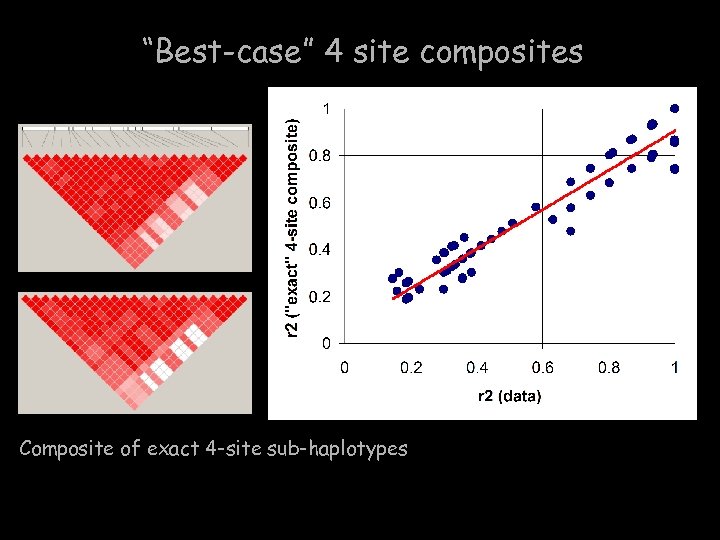 “Best-case” 4 site composites Composite of exact 4 -site sub-haplotypes
“Best-case” 4 site composites Composite of exact 4 -site sub-haplotypes
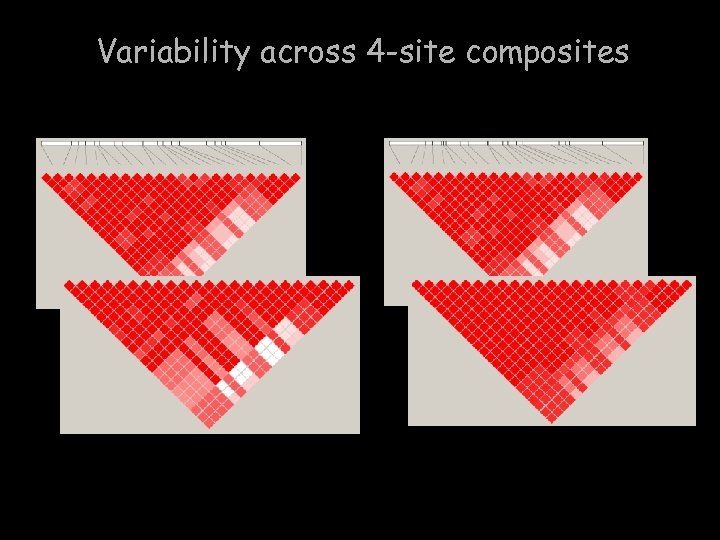 Variability across 4 -site composites
Variability across 4 -site composites
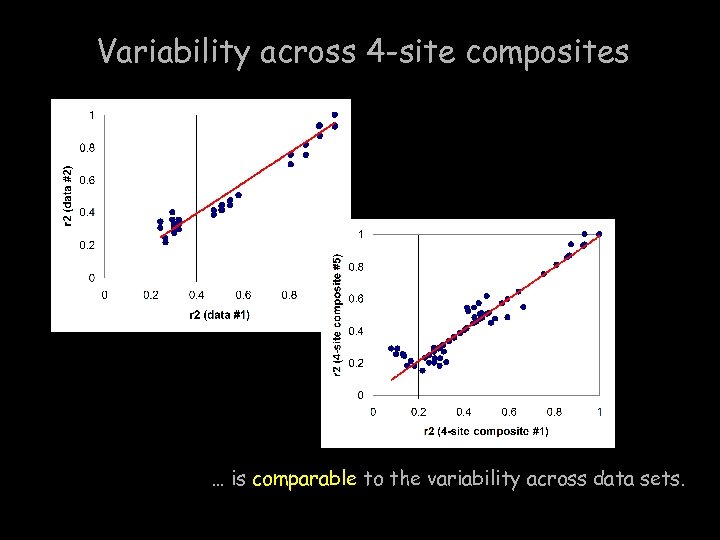 Variability across 4 -site composites … is comparable to the variability across data sets.
Variability across 4 -site composites … is comparable to the variability across data sets.
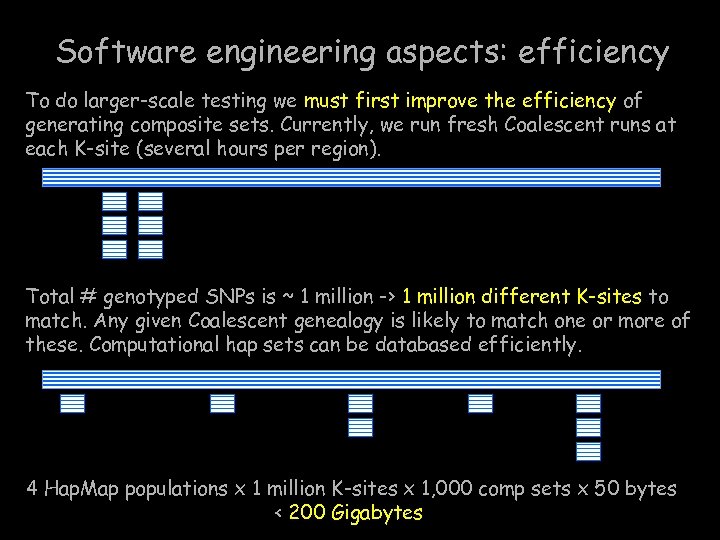 Software engineering aspects: efficiency To do larger-scale testing we must first improve the efficiency of generating composite sets. Currently, we run fresh Coalescent runs at each K-site (several hours per region). Total # genotyped SNPs is ~ 1 million -> 1 million different K-sites to match. Any given Coalescent genealogy is likely to match one or more of these. Computational hap sets can be databased efficiently. 4 Hap. Map populations x 1 million K-sites x 1, 000 comp sets x 50 bytes < 200 Gigabytes
Software engineering aspects: efficiency To do larger-scale testing we must first improve the efficiency of generating composite sets. Currently, we run fresh Coalescent runs at each K-site (several hours per region). Total # genotyped SNPs is ~ 1 million -> 1 million different K-sites to match. Any given Coalescent genealogy is likely to match one or more of these. Computational hap sets can be databased efficiently. 4 Hap. Map populations x 1 million K-sites x 1, 000 comp sets x 50 bytes < 200 Gigabytes
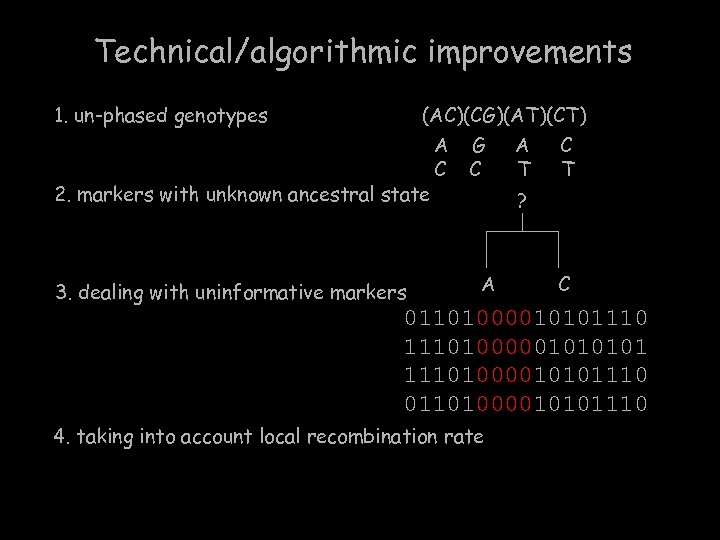 Technical/algorithmic improvements 1. un-phased genotypes (AC)(CG)(AT)(CT) 2. markers with unknown ancestral state 3. dealing with uninformative markers A C G C A T C T ? A C 01101000010101110100000101 11101000010101110 01101000010101110 4. taking into account local recombination rate
Technical/algorithmic improvements 1. un-phased genotypes (AC)(CG)(AT)(CT) 2. markers with unknown ancestral state 3. dealing with uninformative markers A C G C A T C T ? A C 01101000010101110100000101 11101000010101110 01101000010101110 4. taking into account local recombination rate
 Acknowledgements Eric Tsung Aaron Quinlan Ike Unsal Eva Czabarka (Dept. Mathematics, William & Mary)
Acknowledgements Eric Tsung Aaron Quinlan Ike Unsal Eva Czabarka (Dept. Mathematics, William & Mary)


