cd03d80f28e07917dbf0ed60958f26f9.ppt
- Количество слайдов: 26

A. A. 2008 -2009 CORSO DI BIOINFORMATICA per il CLT in Biotecnologie Sanitarie Università di Padova Docente: Dr. STEFANIA BORTOLUZZI

II LEZIONE • Database di interesse per la genetica e la biologia molecolare • Portali per l'accesso a database e servizi bioinformatici
DATABASES AND DATA RETRIEVAL Biosequences and Gene-related info
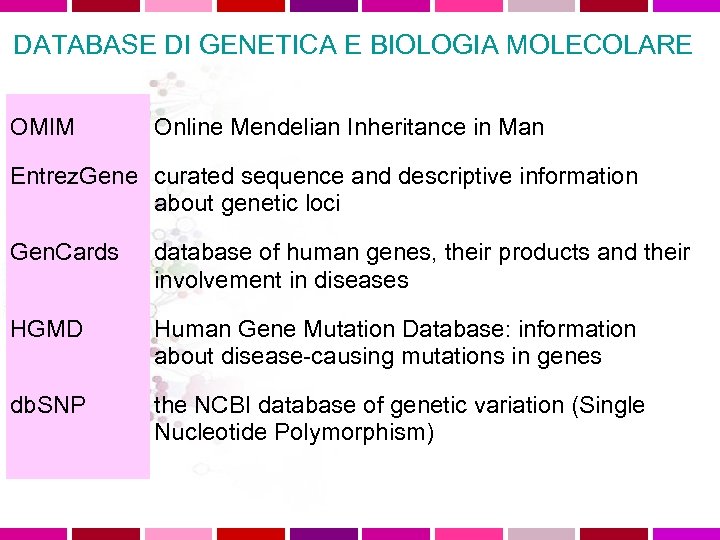
DATABASE DI GENETICA E BIOLOGIA MOLECOLARE OMIM Online Mendelian Inheritance in Man Entrez. Gene curated sequence and descriptive information about genetic loci Gen. Cards database of human genes, their products and their involvement in diseases HGMD Human Gene Mutation Database: information about disease-causing mutations in genes db. SNP the NCBI database of genetic variation (Single Nucleotide Polymorphism)

DATABASE DI GENETICA E BIOLOGIA MOLECOLARE OMIM http: //www. ncbi. nlm. nih. gov/entrez/query. fcgi? db=OMIM Entrez. Gene http: //www. ncbi. nlm. nih. gov/entrez/query. fcgi? CMD=search&DB=gene Gen. Cards http: //bioinformatics. weizmann. ac. il/cards/ HGMD http: //www. hgmd. cf. ac. uk/ac/index. php db. SNP http: //www. ncbi. nlm. nih. gov/projects/SNP/

OMIM Online Mendelian Inheritance in Man Catalogo di geni umani e malattie genetiche (Dr. Victor A. Mc. Kusick, Johns Hopkins + NCBI) Contiene informazione testuale, riferimenti bibliografici e links a MEDLINE, sequenze e ad altre risorse OMIM gene map Posizioni di mappa citogenetica di geni-malattia e altri geni descritti in OMIM morbid map Posizioni di mappa citogenetica di geni-malattia indicizzati in OMIM
Use OMIM to explore Rett syndrome
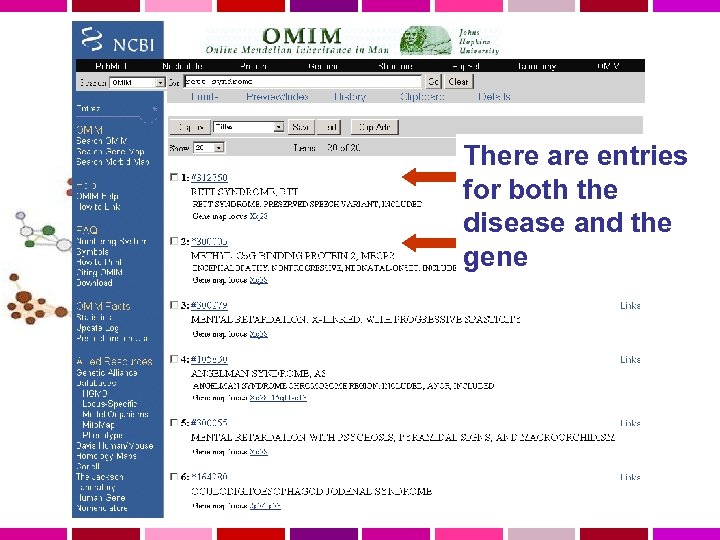
There are entries for both the disease and the gene
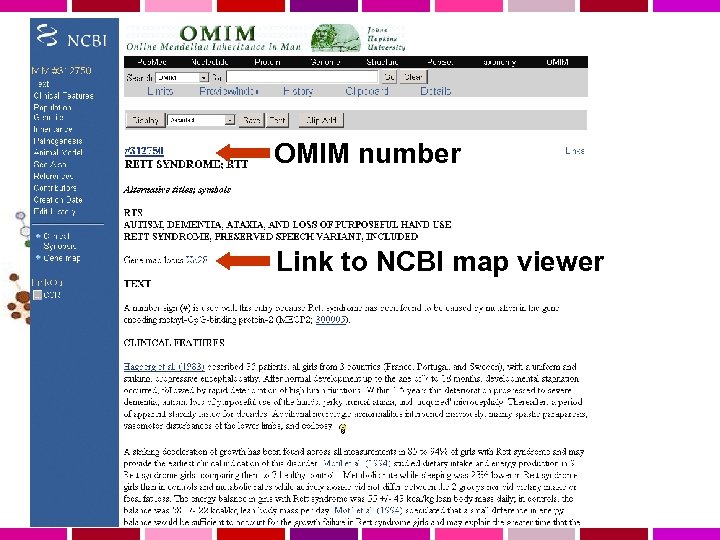
OMIM number Link to NCBI map viewer
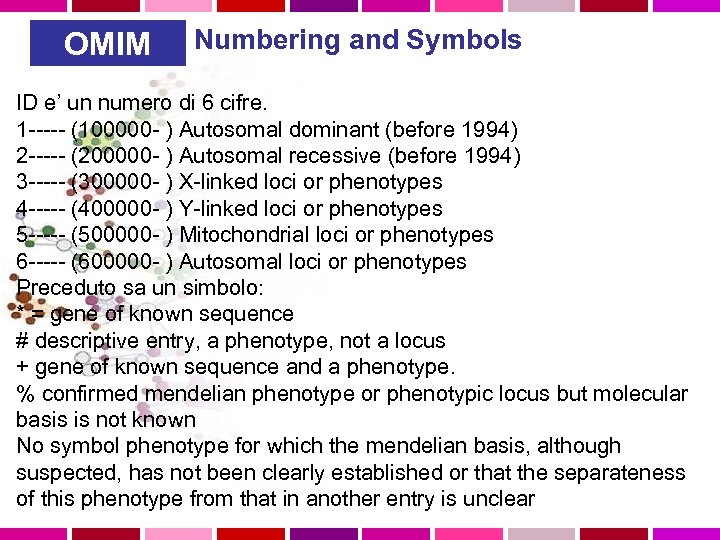
OMIM Numbering and Symbols ID e’ un numero di 6 cifre. 1 ----- (100000 - ) Autosomal dominant (before 1994) 2 ----- (200000 - ) Autosomal recessive (before 1994) 3 ----- (300000 - ) X-linked loci or phenotypes 4 ----- (400000 - ) Y-linked loci or phenotypes 5 ----- (500000 - ) Mitochondrial loci or phenotypes 6 ----- (600000 - ) Autosomal loci or phenotypes Preceduto sa un simbolo: * = gene of known sequence # descriptive entry, a phenotype, not a locus + gene of known sequence and a phenotype. % confirmed mendelian phenotype or phenotypic locus but molecular basis is not known No symbol phenotype for which the mendelian basis, although suspected, has not been clearly established or that the separateness of this phenotype from that in another entry is unclear

OMIM Varianti alleliche stesso ID piu’ un altro numero di 4 cifre. Sono MUTAZIONI CHE CAUSANO MALATTIE
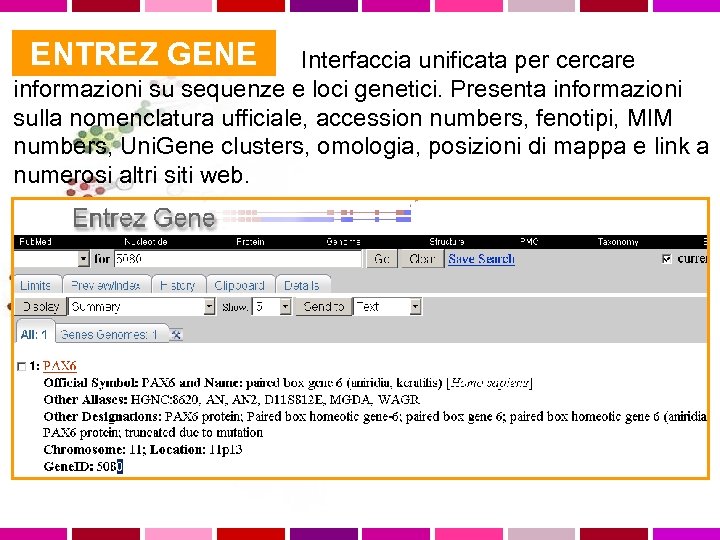
ENTREZ GENE Interfaccia unificata per cercare informazioni su sequenze e loci genetici. Presenta informazioni sulla nomenclatura ufficiale, accession numbers, fenotipi, MIM numbers, Uni. Gene clusters, omologia, posizioni di mappa e link a numerosi altri siti web.

ENTREZ GENE
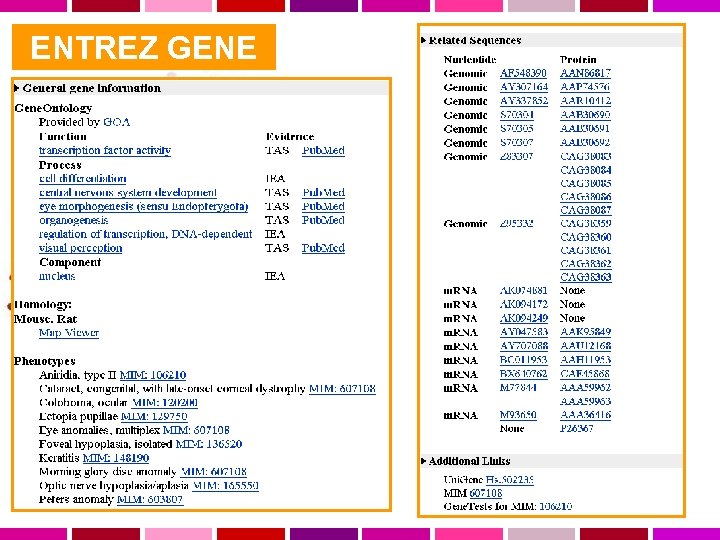
ENTREZ GENE
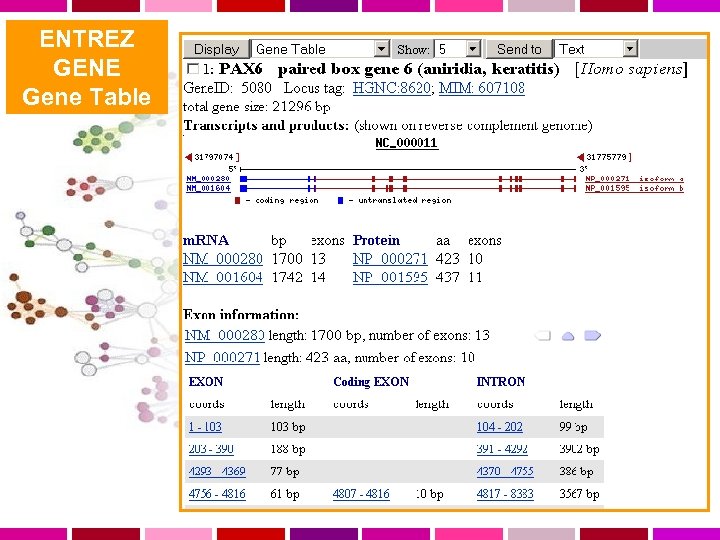
ENTREZ GENE Gene Table

Gene. Cards Weizmann Institute of Science, Israele Gene. Cards is a database of human genes, their products and their involvement in diseases. It offers concise information about the functions of all human genes that have an approved symbol, as well as selected others. The information presented here has been automatically extracted from various resources. Gene. Cards™ is particulary useful for people who wish to find information about genes of interest in the context of functional genomics and proteomics. http: //www. genecards. org/
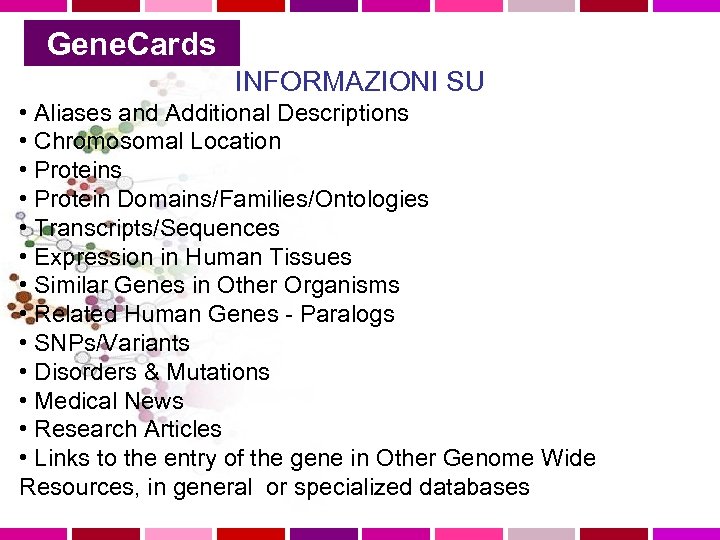
Gene. Cards INFORMAZIONI SU • Aliases and Additional Descriptions • Chromosomal Location • Proteins • Protein Domains/Families/Ontologies • Transcripts/Sequences • Expression in Human Tissues • Similar Genes in Other Organisms • Related Human Genes - Paralogs • SNPs/Variants • Disorders & Mutations • Medical News • Research Articles • Links to the entry of the gene in Other Genome Wide Resources, in general or specialized databases
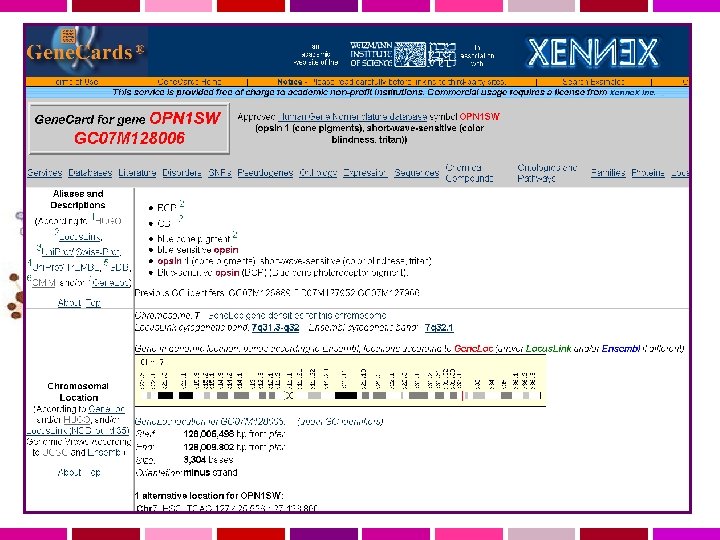
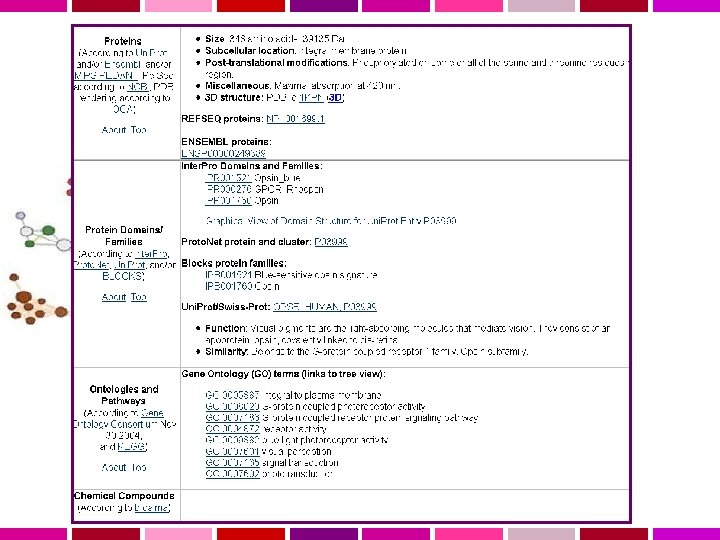


Gene. Cards is an integrated database of human genes that includes automatically-mined genomic, proteomic and transcriptomic information, as well as orthologies, disease relationships, SNPs, gene expression, gene function and more. Gene. Note is a database of human genes and their expression profiles in healthy tissues (Weizmann Institute of Science DNA array experiments, Affymetrix HG-U 95 ) Gene. Annot provides a revised and improved annotation of Affymetrix probe-sets from HG-U 95, HG-U 133 and HG-U 133 Plus 2. 0. Gene. Loc presents an integrated map for each human chromosome, based on data integrated by the Gene. Loc algorithm. Gene. Loc includes further links to Gene. Cards, NCBI's Human Genome Sequencing, Uni. Gene, Genome Database, and mapping resources. Gene. Tide is an automated system for human transcripts (m. RNA & ESTs) annotation and elucidation of de-novo genes.

HGMD http: //www. hgmd. cf. ac. uk/ac/index. php • Human Gene Mutation Database (HGMD) raccoglie le mutazioni conosciute (pubblicate) in geni umani, responsabili di malattie genetiche • Creato per studiare il meccanismo delle mutazioni nel genoma umano, per riconoscere le regioni e i loci ipermutabili • Ora e’ importante anche come raccolta di dati. Utile per diagnosi molecolare di patologie e consulenza genetica. • Non include mutazioni somatiche o mitocondriali, mutazioni silenti. • Dal marzo 1999, HGMD include disease-associated polymorphisms. • Basato sull’analisi di >250 riviste scientifiche.
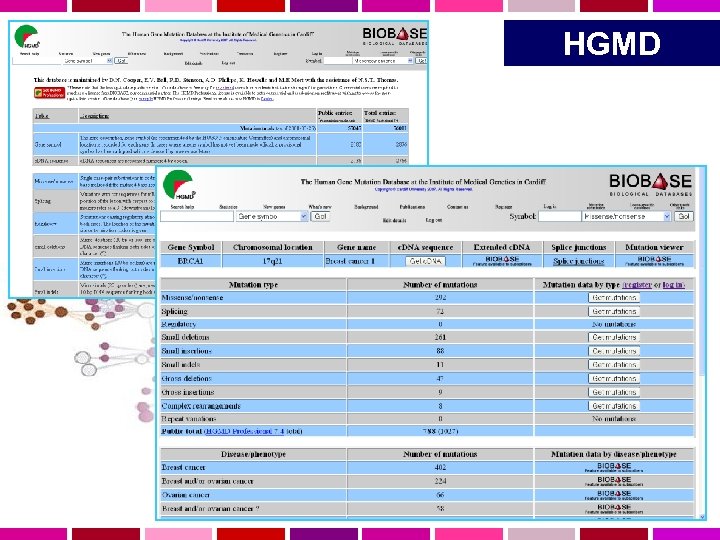
HGMD
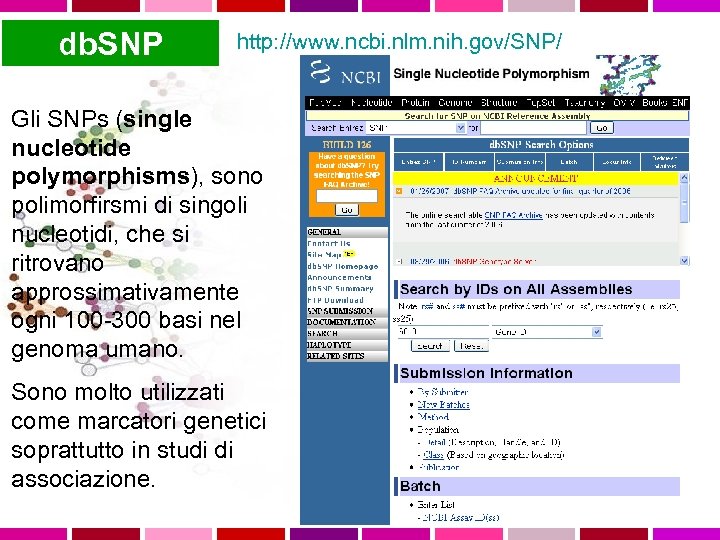
db. SNP http: //www. ncbi. nlm. nih. gov/SNP/ Gli SNPs (single nucleotide polymorphisms), sono polimorfirsmi di singoli nucleotidi, che si ritrovano approssimativamente ogni 100 -300 basi nel genoma umano. Sono molto utilizzati come marcatori genetici soprattutto in studi di associazione.
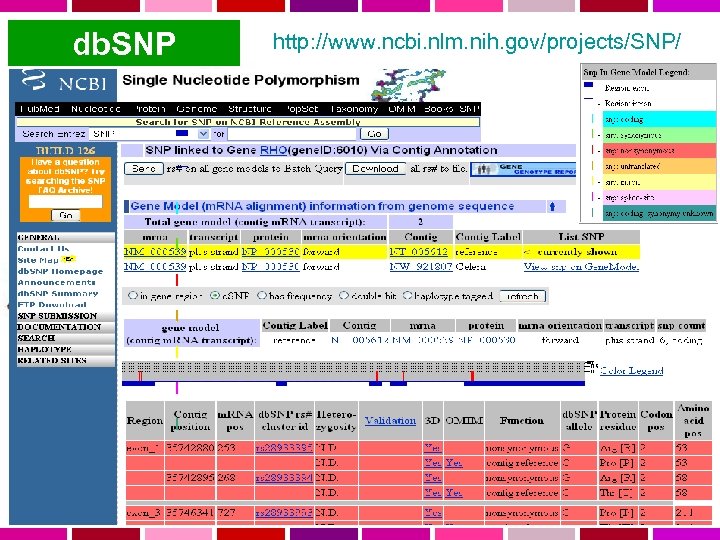
db. SNP http: //www. ncbi. nlm. nih. gov/projects/SNP/
Portali per l'accesso a database e servizi bioinformatici NCBI (SNP, SAGE, Gene Expression Omnibus, Cancer Chromosomes database, …) Ex. PASy Proteomics Server (http: //www. expasy. org/) EBI (EMBL Nucleotide Database, Uni. Prot Knowledgebase, Macromol. Structure Database, Array. Express, Ensembl, …, TOOLBOX) (http: //www. ebi. ac. uk/services/)
cd03d80f28e07917dbf0ed60958f26f9.ppt