4e85c2344eaaa1b1abdbe4a535f7c1a3.ppt
- Количество слайдов: 22
 9 th International Conference on Intelligent System for Molecular Biology Tivoli Gardens, Copenhagen, Denmark July 19 -26, 2001 Park, Ji-Yoon
9 th International Conference on Intelligent System for Molecular Biology Tivoli Gardens, Copenhagen, Denmark July 19 -26, 2001 Park, Ji-Yoon
 Satellite Meetings(July 19 -20) July 19 - The Open Source author’s contract : Steven Brenner - Bio. Java project report: Thomas Down and Matthew Pocock - Biopython project report: Andrew Dalke - Bio. CORBA project report: Jason Stajich - biok: Catherine Letondal - Lightning Talks: • OMG’s new Model Driven Architecture(MDA): Scott Markel • Bio. Ruby: Yoshinori Okuju, Toshiaki Katayama, and Mitsuteru Nakao • Genquire: David Block • Generation and use of substitution matrices in Biopython: IDDo Friedberg and Brad Chapman
Satellite Meetings(July 19 -20) July 19 - The Open Source author’s contract : Steven Brenner - Bio. Java project report: Thomas Down and Matthew Pocock - Biopython project report: Andrew Dalke - Bio. CORBA project report: Jason Stajich - biok: Catherine Letondal - Lightning Talks: • OMG’s new Model Driven Architecture(MDA): Scott Markel • Bio. Ruby: Yoshinori Okuju, Toshiaki Katayama, and Mitsuteru Nakao • Genquire: David Block • Generation and use of substitution matrices in Biopython: IDDo Friedberg and Brad Chapman
 Satellite Meetings(July 19 -20) July 20 : Biopathway - Bioperl project report: Hilmar Lapp - Ens. EMBL project report: Arne Stabenau - Lightning Talks: • Bioperl-project: Ewan Birney • Open. BSA: Juha Muilu, Martin Senger and Alan Robinson • Genetic algorithm and neural network libraries: Brad Chapman • Mining gene expression information using a controlled hierarchical vocabulary : Peter van heusden • TFBS: Perl modules for transcription factor detection and analysis: Boris Lenhard - A tool suite for the Gene Ontology: Chris Mungall, Hohn Richter, Bradley Marshall, and Suzanna Lewis - De. CAL: A system for constructing comparative maps: Debra Goldberg, Jon Kleinberg, and Susan Mc. Couch
Satellite Meetings(July 19 -20) July 20 : Biopathway - Bioperl project report: Hilmar Lapp - Ens. EMBL project report: Arne Stabenau - Lightning Talks: • Bioperl-project: Ewan Birney • Open. BSA: Juha Muilu, Martin Senger and Alan Robinson • Genetic algorithm and neural network libraries: Brad Chapman • Mining gene expression information using a controlled hierarchical vocabulary : Peter van heusden • TFBS: Perl modules for transcription factor detection and analysis: Boris Lenhard - A tool suite for the Gene Ontology: Chris Mungall, Hohn Richter, Bradley Marshall, and Suzanna Lewis - De. CAL: A system for constructing comparative maps: Debra Goldberg, Jon Kleinberg, and Susan Mc. Couch
 Tutorial( July 21) * Morning Turorials: Sat July 21 8: 30 -12: 30 [Statistical analysis of micro-arrays studies] : Emmanuel Lazaridis, University of South Florida Afternoon Turorials: Sat July 21 14: 00 -18: 00 [Network genomics] : Christian Forst, Los Alamos National Laboratory
Tutorial( July 21) * Morning Turorials: Sat July 21 8: 30 -12: 30 [Statistical analysis of micro-arrays studies] : Emmanuel Lazaridis, University of South Florida Afternoon Turorials: Sat July 21 14: 00 -18: 00 [Network genomics] : Christian Forst, Los Alamos National Laboratory
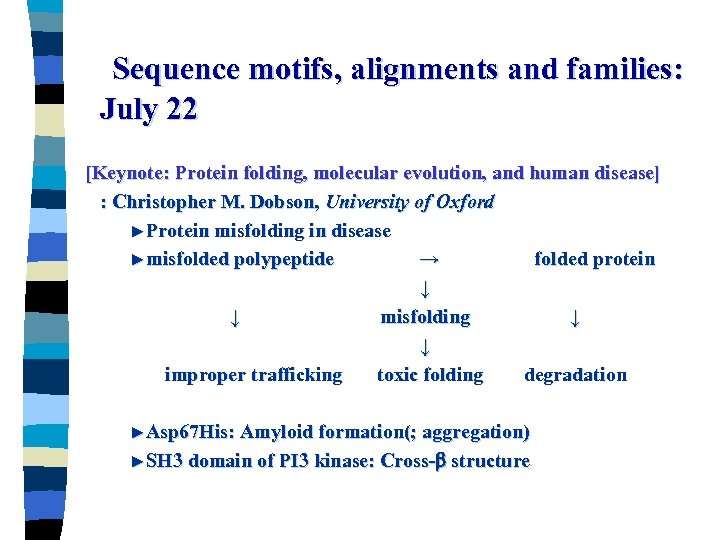 Sequence motifs, alignments and families: July 22 [Keynote: Protein folding, molecular evolution, and human disease] : Christopher M. Dobson, University of Oxford ►Protein misfolding in disease ►misfolded polypeptide → folded protein ↓ ↓ misfolding ↓ ↓ improper trafficking toxic folding degradation ►Asp 67 His: Amyloid formation(; aggregation) ►SH 3 domain of PI 3 kinase: Cross- structure
Sequence motifs, alignments and families: July 22 [Keynote: Protein folding, molecular evolution, and human disease] : Christopher M. Dobson, University of Oxford ►Protein misfolding in disease ►misfolded polypeptide → folded protein ↓ ↓ misfolding ↓ ↓ improper trafficking toxic folding degradation ►Asp 67 His: Amyloid formation(; aggregation) ►SH 3 domain of PI 3 kinase: Cross- structure
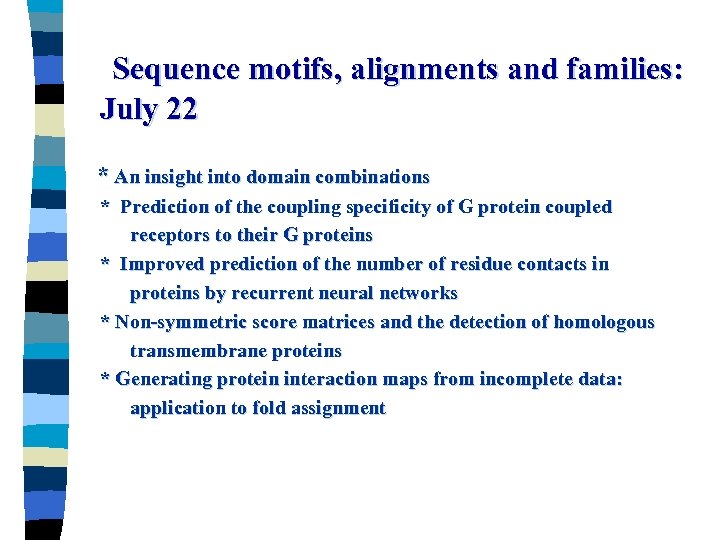 Sequence motifs, alignments and families: July 22 * An insight into domain combinations * Prediction of the coupling specificity of G protein coupled receptors to their G proteins * Improved prediction of the number of residue contacts in proteins by recurrent neural networks * Non-symmetric score matrices and the detection of homologous transmembrane proteins * Generating protein interaction maps from incomplete data: application to fold assignment
Sequence motifs, alignments and families: July 22 * An insight into domain combinations * Prediction of the coupling specificity of G protein coupled receptors to their G proteins * Improved prediction of the number of residue contacts in proteins by recurrent neural networks * Non-symmetric score matrices and the detection of homologous transmembrane proteins * Generating protein interaction maps from incomplete data: application to fold assignment
![Sequence motifs, alignments and families: July 22 [Keynote - Structural Genomics] - Christopher M. Sequence motifs, alignments and families: July 22 [Keynote - Structural Genomics] - Christopher M.](https://present5.com/presentation/4e85c2344eaaa1b1abdbe4a535f7c1a3/image-7.jpg) Sequence motifs, alignments and families: July 22 [Keynote - Structural Genomics] - Christopher M. Dobson, University of Oxford - Goal : All protein domains carry all functional families How many experimental structure? - Coordination of international programs in structural genomics - Pathways in expression profile * 0 j-py: a software tool for low complexity proteins and protein domains : Michael J. Wise, Centre for Communications Systems Research → new tool for looking peptide * Separating real motifs from their artifacts * Feature selection for DNA methylation based cancer classification * An algorithm for finding signals of unknown length in DNA sequences
Sequence motifs, alignments and families: July 22 [Keynote - Structural Genomics] - Christopher M. Dobson, University of Oxford - Goal : All protein domains carry all functional families How many experimental structure? - Coordination of international programs in structural genomics - Pathways in expression profile * 0 j-py: a software tool for low complexity proteins and protein domains : Michael J. Wise, Centre for Communications Systems Research → new tool for looking peptide * Separating real motifs from their artifacts * Feature selection for DNA methylation based cancer classification * An algorithm for finding signals of unknown length in DNA sequences
![Networks and Modeling: July 23 [Keynote- Protein Interactions] : David Eisenberg, University of California, Networks and Modeling: July 23 [Keynote- Protein Interactions] : David Eisenberg, University of California,](https://present5.com/presentation/4e85c2344eaaa1b1abdbe4a535f7c1a3/image-8.jpg) Networks and Modeling: July 23 [Keynote- Protein Interactions] : David Eisenberg, University of California, Los. Angeles • Rossetta Stone - Fusion of functionally-linked domain - http: // dip. doe-mbi. ucla. edu • Phylogenic profile - correlated occurrence of pairs of proteins in genomes • Gene function • Database interacting protein • 3 D domain swapping • Signaling path
Networks and Modeling: July 23 [Keynote- Protein Interactions] : David Eisenberg, University of California, Los. Angeles • Rossetta Stone - Fusion of functionally-linked domain - http: // dip. doe-mbi. ucla. edu • Phylogenic profile - correlated occurrence of pairs of proteins in genomes • Gene function • Database interacting protein • 3 D domain swapping • Signaling path
 Networks and Modeling: July 23
Networks and Modeling: July 23
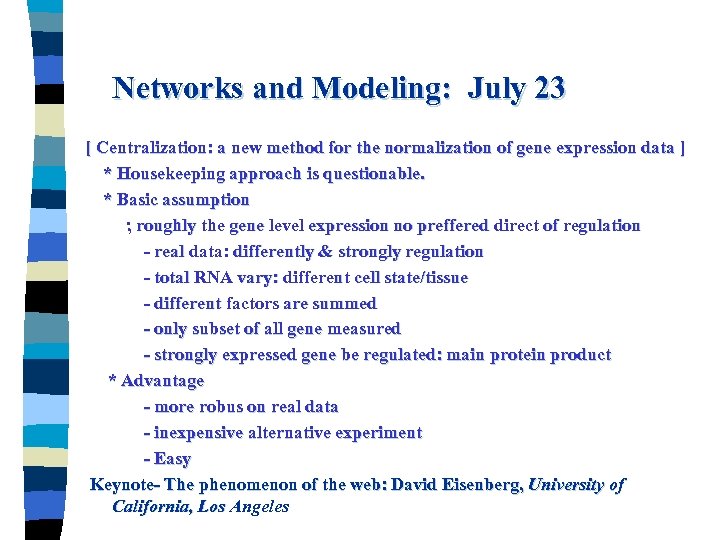 Networks and Modeling: July 23 [ Centralization: a new method for the normalization of gene expression data ] * Housekeeping approach is questionable. * Basic assumption ; roughly the gene level expression no preffered direct of regulation - real data: differently & strongly regulation - total RNA vary: different cell state/tissue - different factors are summed - only subset of all gene measured - strongly expressed gene be regulated: main protein product * Advantage - more robus on real data - inexpensive alternative experiment - Easy Keynote- The phenomenon of the web: David Eisenberg, University of California, Los Angeles
Networks and Modeling: July 23 [ Centralization: a new method for the normalization of gene expression data ] * Housekeeping approach is questionable. * Basic assumption ; roughly the gene level expression no preffered direct of regulation - real data: differently & strongly regulation - total RNA vary: different cell state/tissue - different factors are summed - only subset of all gene measured - strongly expressed gene be regulated: main protein product * Advantage - more robus on real data - inexpensive alternative experiment - Easy Keynote- The phenomenon of the web: David Eisenberg, University of California, Los Angeles
![Networks and Modeling: July 23 [Keynote- The phenomenon of the web] : David Eisenberg, Networks and Modeling: July 23 [Keynote- The phenomenon of the web] : David Eisenberg,](https://present5.com/presentation/4e85c2344eaaa1b1abdbe4a535f7c1a3/image-11.jpg) Networks and Modeling: July 23 [Keynote- The phenomenon of the web] : David Eisenberg, University of California, Los Angeles http: // rana. lbl. gov http: // genome-www. standford. edu. /microarray
Networks and Modeling: July 23 [Keynote- The phenomenon of the web] : David Eisenberg, University of California, Los Angeles http: // rana. lbl. gov http: // genome-www. standford. edu. /microarray
 Gene structure, Regulation, and Modeling: July 24 * Keynote- The Modern RNA world: many genes don’t encode proteins : Sean Eddy, Washington University * Promoter prediction in the human genome * Joint modeling of DNA sequence and physical properties to improve eukaryotic promoter recognition * Computational expansion of genetic networks * GENIES: a natural-language processing systems for the extraction of molecular pathways from journal articles * Designing better phages * Computational Analysis of RNA splicing * Disambiguating proteins, genes, and RNA in text: a machine learning approach * Gene recognition based on DAG shortest paths * An efficient algorithm for finding short approximate non-tandem repeats
Gene structure, Regulation, and Modeling: July 24 * Keynote- The Modern RNA world: many genes don’t encode proteins : Sean Eddy, Washington University * Promoter prediction in the human genome * Joint modeling of DNA sequence and physical properties to improve eukaryotic promoter recognition * Computational expansion of genetic networks * GENIES: a natural-language processing systems for the extraction of molecular pathways from journal articles * Designing better phages * Computational Analysis of RNA splicing * Disambiguating proteins, genes, and RNA in text: a machine learning approach * Gene recognition based on DAG shortest paths * An efficient algorithm for finding short approximate non-tandem repeats
 Methods: July 25 * Keynote- Membrane proteins: From the computer to the bench and back: Gunnar von Heijne, Stockholm University * Design of a compartmentalized shotgun assembler for the human genome * Probabilistic approaches to the use of higher order clone relationships in physical map assembly: * Fragment assembly with double-barreled data * SCOPE: a probabilistic model for scoring tandem mass spectra against a peptide database * Probe selection algorithms with applications in the analysis of microbial communities * Fast optimal leaf ordering for hierarchical clustering * Separation of samples into their constituents using gene expression data
Methods: July 25 * Keynote- Membrane proteins: From the computer to the bench and back: Gunnar von Heijne, Stockholm University * Design of a compartmentalized shotgun assembler for the human genome * Probabilistic approaches to the use of higher order clone relationships in physical map assembly: * Fragment assembly with double-barreled data * SCOPE: a probabilistic model for scoring tandem mass spectra against a peptide database * Probe selection algorithms with applications in the analysis of microbial communities * Fast optimal leaf ordering for hierarchical clustering * Separation of samples into their constituents using gene expression data
 WEB: July 26 * Education in Bioinformatics: Current Trends and Issues. - Shoba Ranganathan * Opening Addresss: Bioinformatics Education - Looking to the Future: Russ Altman * The S* Life Science Informatics Alliance - Shoba Ranganathan * Bioinformatics BS at the Univerisity of California, Santa Cruz - Kevin Karplus * A Masters Degree in Bioinformatics in Switzerland - Patricia Palagi * Emerging US & UK Standards for Graduate Bioinformatics training - Linda Ellis
WEB: July 26 * Education in Bioinformatics: Current Trends and Issues. - Shoba Ranganathan * Opening Addresss: Bioinformatics Education - Looking to the Future: Russ Altman * The S* Life Science Informatics Alliance - Shoba Ranganathan * Bioinformatics BS at the Univerisity of California, Santa Cruz - Kevin Karplus * A Masters Degree in Bioinformatics in Switzerland - Patricia Palagi * Emerging US & UK Standards for Graduate Bioinformatics training - Linda Ellis
 WEB: July 26 * Bioinformatics Course Delivery: Tools and Infrastructure. Siv Andersson(Chair) * Bioinformatics: Introducing the concept of “ evaluation-based” learning : Siv Andersson * Problem-oriented sequence analysis tool: Ueng-Cheng Yang * EMBER- A European Multimedia Bioinformatics Educational Resource: C. Victor Jongeneel * Virtual Reality and Visualization for Bioinformatics Education: YY Cai * Starting a new Bioinformatics Program. Phyllis Gardner(Chair) * Initiating a multi-disciplinary, trans-institutional program: A Dean’s perspective: Phyllis Gardner * Insights into starting a new Multi-disciplinary program: Betty Cheng
WEB: July 26 * Bioinformatics Course Delivery: Tools and Infrastructure. Siv Andersson(Chair) * Bioinformatics: Introducing the concept of “ evaluation-based” learning : Siv Andersson * Problem-oriented sequence analysis tool: Ueng-Cheng Yang * EMBER- A European Multimedia Bioinformatics Educational Resource: C. Victor Jongeneel * Virtual Reality and Visualization for Bioinformatics Education: YY Cai * Starting a new Bioinformatics Program. Phyllis Gardner(Chair) * Initiating a multi-disciplinary, trans-institutional program: A Dean’s perspective: Phyllis Gardner * Insights into starting a new Multi-disciplinary program: Betty Cheng
 WEB: July 26 * Bioinformatics Training. Frederique Galisson(Chair) * The Canadian Bioinformatics Workshops: Stephen Herst * The Bio. Navigator Education Package- resources for practical instruction in bioinformatics: Bruno A. Gaeta * The Human Genome Mapping Project Resources Centre. Encouraging Bioinformatics Awareness: Lisa Mullan * Panel Discussion: Betty Cheng(Chair) The S* Life Science Informatics Alliance: Question Time: S* Team * Concluding Session: Closing Remarks. Shoba Ranganathan(Chair) Bioinformatics Education: Future trends and perspectives: Philip Bourne
WEB: July 26 * Bioinformatics Training. Frederique Galisson(Chair) * The Canadian Bioinformatics Workshops: Stephen Herst * The Bio. Navigator Education Package- resources for practical instruction in bioinformatics: Bruno A. Gaeta * The Human Genome Mapping Project Resources Centre. Encouraging Bioinformatics Awareness: Lisa Mullan * Panel Discussion: Betty Cheng(Chair) The S* Life Science Informatics Alliance: Question Time: S* Team * Concluding Session: Closing Remarks. Shoba Ranganathan(Chair) Bioinformatics Education: Future trends and perspectives: Philip Bourne

 Free Energy( ∆ G ) Thermodynamic constant that gives the amount of energy required for or released by a reaction - kcal/mol - Reaction that require energy ; positive - Reation that release free energy ; negative - Energy must be released overall to form a base-paired structure - The stability of the structure is determined by the amount of energy released
Free Energy( ∆ G ) Thermodynamic constant that gives the amount of energy required for or released by a reaction - kcal/mol - Reaction that require energy ; positive - Reation that release free energy ; negative - Energy must be released overall to form a base-paired structure - The stability of the structure is determined by the amount of energy released
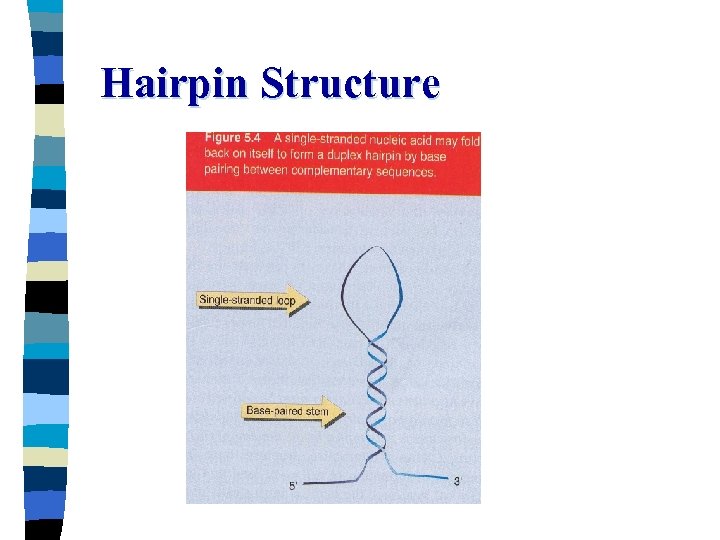 Hairpin Structure
Hairpin Structure
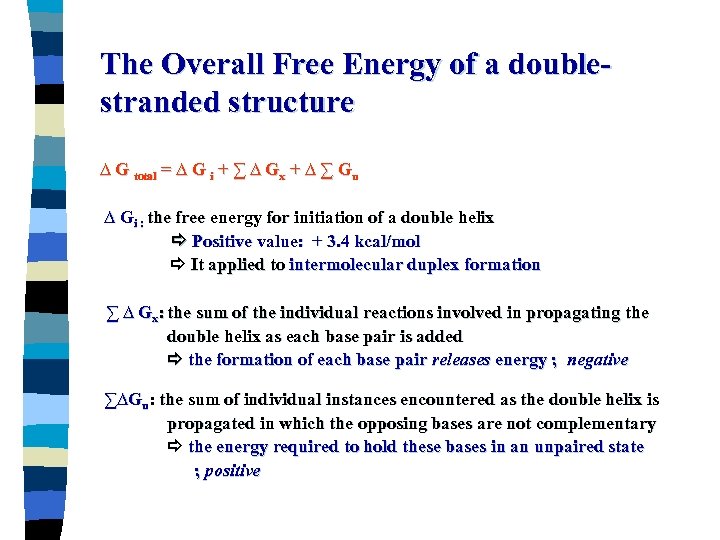 The Overall Free Energy of a doublestranded structure ∆ G total = ∆ G i + ∑ ∆ Gx + ∆ ∑ Gu ∆ Gi : the free energy for initiation of a double helix Positive value: + 3. 4 kcal/mol It applied to intermolecular duplex formation ∑ ∆ Gx: the sum of the individual reactions involved in propagating the double helix as each base pair is added the formation of each base pair releases energy ; negative ∑∆Gu: the sum of individual instances encountered as the double helix is propagated in which the opposing bases are not complementary the energy required to hold these bases in an unpaired state ; positive
The Overall Free Energy of a doublestranded structure ∆ G total = ∆ G i + ∑ ∆ Gx + ∆ ∑ Gu ∆ Gi : the free energy for initiation of a double helix Positive value: + 3. 4 kcal/mol It applied to intermolecular duplex formation ∑ ∆ Gx: the sum of the individual reactions involved in propagating the double helix as each base pair is added the formation of each base pair releases energy ; negative ∑∆Gu: the sum of individual instances encountered as the double helix is propagated in which the opposing bases are not complementary the energy required to hold these bases in an unpaired state ; positive
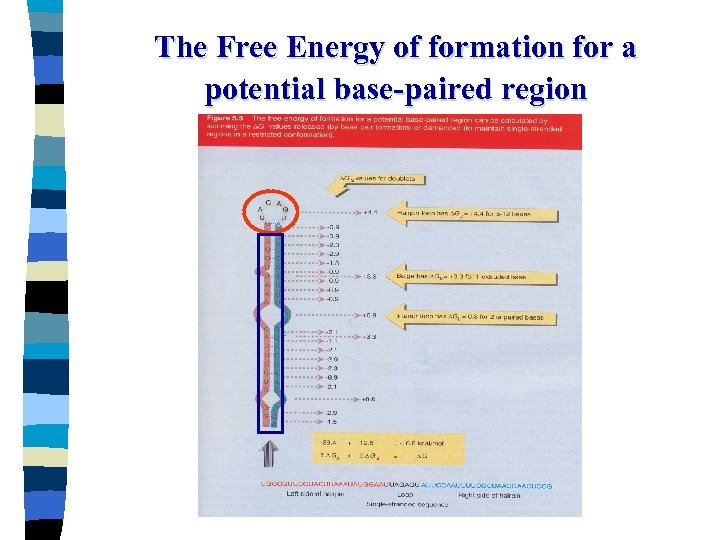 The Free Energy of formation for a potential base-paired region
The Free Energy of formation for a potential base-paired region
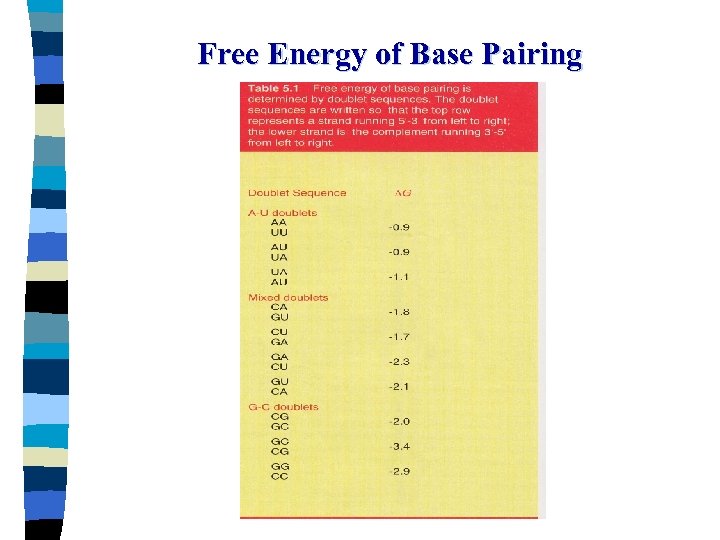 Free Energy of Base Pairing
Free Energy of Base Pairing


