796d0433e0d909967b18d4dbf336b78f.ppt
- Количество слайдов: 55
 3/19/2018 Master title Molecular Interactions – the Int. Act Database Sandra Orchard EMBL-EBI is an Outstation of the European Molecular Biology Laboratory.
3/19/2018 Master title Molecular Interactions – the Int. Act Database Sandra Orchard EMBL-EBI is an Outstation of the European Molecular Biology Laboratory.
 Why are we interested in Interactions ? 1. As a means of precisely understanding a protein role inside a specific cell type 2. To verify data, visualise your own interaction netwrok over the known space 3. Guilt by Association – it may be the only means of predicting a protein’s function 4. As building blocks for System’s Biology and Drug Discovery What data are we dealing with ?
Why are we interested in Interactions ? 1. As a means of precisely understanding a protein role inside a specific cell type 2. To verify data, visualise your own interaction netwrok over the known space 3. Guilt by Association – it may be the only means of predicting a protein’s function 4. As building blocks for System’s Biology and Drug Discovery What data are we dealing with ?
 Why are there so many issues with interaction data? 1. Wide variety of methods for demonstrating molecular interactions – all have their strengths and weaknesses 2. No single method accurately defines an interaction as being a true binary interaction observed under physiological conditions
Why are there so many issues with interaction data? 1. Wide variety of methods for demonstrating molecular interactions – all have their strengths and weaknesses 2. No single method accurately defines an interaction as being a true binary interaction observed under physiological conditions
 Why do we need interaction databases • Issues with all interaction data – true picture can only be built up by combining data derived using multiple techniques, multiple laboratories • Problematic for any bench researcher to do – issues with data formats, molecular identifiers, sheer volume of data • Molecular interaction databases publicly funded to collect this data and annotate in a format most useful to researchers
Why do we need interaction databases • Issues with all interaction data – true picture can only be built up by combining data derived using multiple techniques, multiple laboratories • Problematic for any bench researcher to do – issues with data formats, molecular identifiers, sheer volume of data • Molecular interaction databases publicly funded to collect this data and annotate in a format most useful to researchers
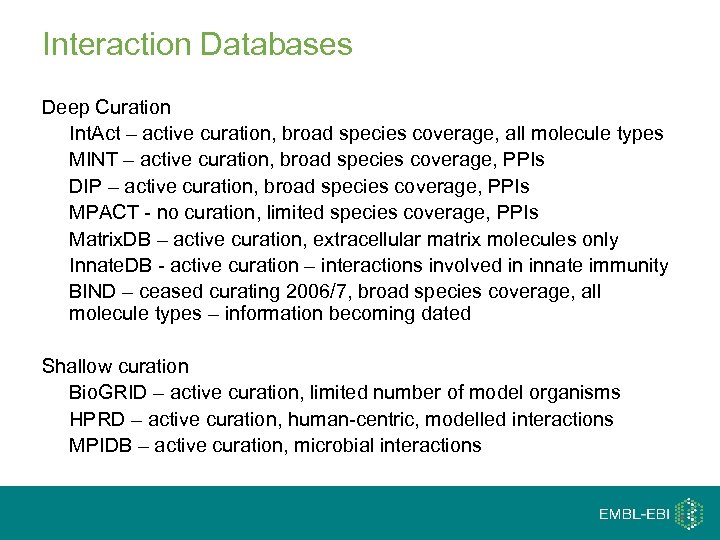 Interaction Databases Deep Curation Int. Act – active curation, broad species coverage, all molecule types MINT – active curation, broad species coverage, PPIs DIP – active curation, broad species coverage, PPIs MPACT - no curation, limited species coverage, PPIs Matrix. DB – active curation, extracellular matrix molecules only Innate. DB - active curation – interactions involved in innate immunity BIND – ceased curating 2006/7, broad species coverage, all molecule types – information becoming dated Shallow curation Bio. GRID – active curation, limited number of model organisms HPRD – active curation, human-centric, modelled interactions MPIDB – active curation, microbial interactions
Interaction Databases Deep Curation Int. Act – active curation, broad species coverage, all molecule types MINT – active curation, broad species coverage, PPIs DIP – active curation, broad species coverage, PPIs MPACT - no curation, limited species coverage, PPIs Matrix. DB – active curation, extracellular matrix molecules only Innate. DB - active curation – interactions involved in innate immunity BIND – ceased curating 2006/7, broad species coverage, all molecule types – information becoming dated Shallow curation Bio. GRID – active curation, limited number of model organisms HPRD – active curation, human-centric, modelled interactions MPIDB – active curation, microbial interactions
 Int. Act goals & achievements 1. Publicly available repository of molecular interactions (mainly PPIs) - >250 K binary interactions taken from >4, 700 publications 2. Data is standards-compliant and available via our website, for download at our ftp site or via PSICQUIC http: //www. ebi. ac. uk/intact ftp: //ftp. ebi. ac. uk/pub/databases/intact www. ebi. ac. uk/Tools/webservices/psicquic/view/main. xhtml 3. Provide open-access versions of the software to allow installation of local Int. Act nodes. 6
Int. Act goals & achievements 1. Publicly available repository of molecular interactions (mainly PPIs) - >250 K binary interactions taken from >4, 700 publications 2. Data is standards-compliant and available via our website, for download at our ftp site or via PSICQUIC http: //www. ebi. ac. uk/intact ftp: //ftp. ebi. ac. uk/pub/databases/intact www. ebi. ac. uk/Tools/webservices/psicquic/view/main. xhtml 3. Provide open-access versions of the software to allow installation of local Int. Act nodes. 6
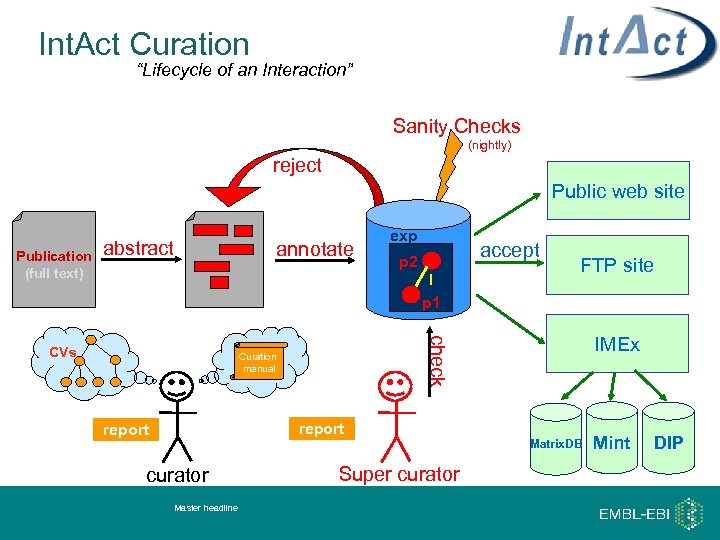 Int. Act Curation “Lifecycle of an Interaction” Sanity Checks (nightly) reject Public web site Publication (full text) abstract . exp accept p 2 I p 1 Curation manual report curator Master headline FTP site IMEx check CVs annotate Super curator Matrix. DB Mint DIP
Int. Act Curation “Lifecycle of an Interaction” Sanity Checks (nightly) reject Public web site Publication (full text) abstract . exp accept p 2 I p 1 Curation manual report curator Master headline FTP site IMEx check CVs annotate Super curator Matrix. DB Mint DIP
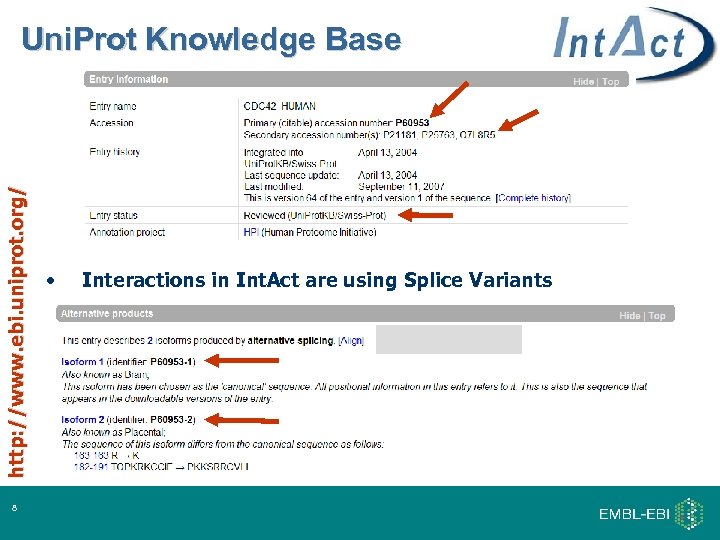 http: //www. ebi. uniprot. org/ Uni. Prot Knowledge Base 8 • Interactions in Int. Act are using Splice Variants
http: //www. ebi. uniprot. org/ Uni. Prot Knowledge Base 8 • Interactions in Int. Act are using Splice Variants
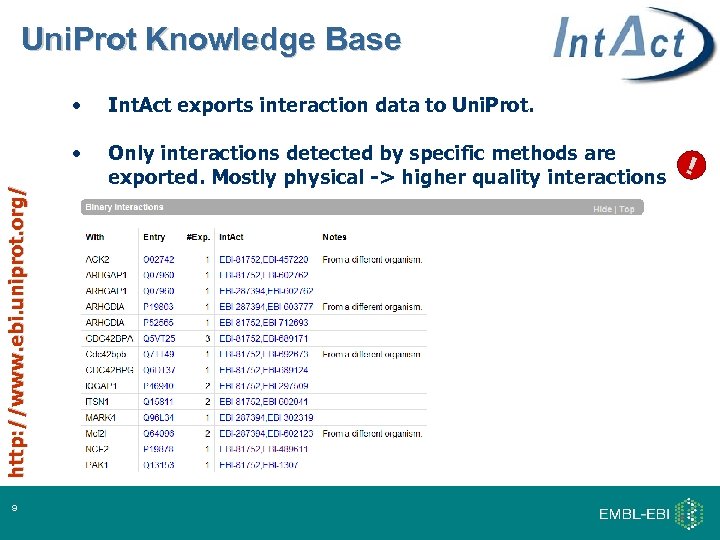 Uni. Prot Knowledge Base 9 Int. Act exports interaction data to Uni. Prot. • http: //www. ebi. uniprot. org/ • Only interactions detected by specific methods are exported. Mostly physical -> higher quality interactions !
Uni. Prot Knowledge Base 9 Int. Act exports interaction data to Uni. Prot. • http: //www. ebi. uniprot. org/ • Only interactions detected by specific methods are exported. Mostly physical -> higher quality interactions !
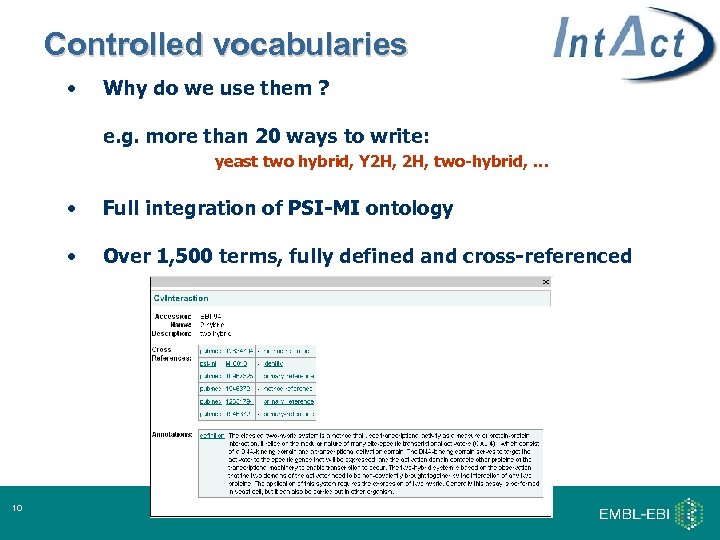 Controlled vocabularies • Why do we use them ? e. g. more than 20 ways to write: yeast two hybrid, Y 2 H, two-hybrid, … • • 10 Full integration of PSI-MI ontology Over 1, 500 terms, fully defined and cross-referenced
Controlled vocabularies • Why do we use them ? e. g. more than 20 ways to write: yeast two hybrid, Y 2 H, two-hybrid, … • • 10 Full integration of PSI-MI ontology Over 1, 500 terms, fully defined and cross-referenced
 Controlled vocabularies www. ebi. ac. uk/ols
Controlled vocabularies www. ebi. ac. uk/ols
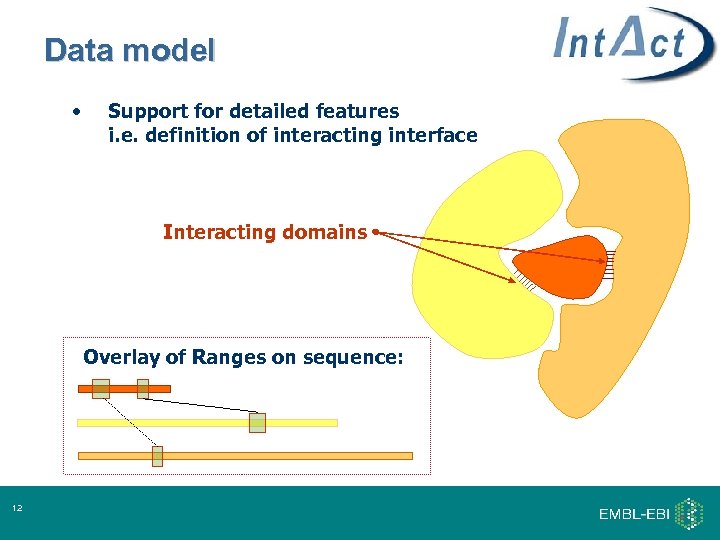 Data model • Support for detailed features i. e. definition of interacting interface Interacting domains Overlay of Ranges on sequence: 12
Data model • Support for detailed features i. e. definition of interacting interface Interacting domains Overlay of Ranges on sequence: 12
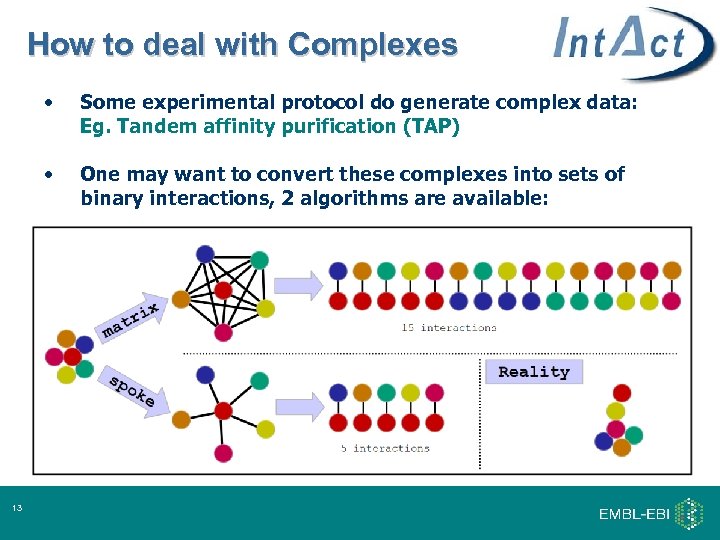 How to deal with Complexes • • 13 Some experimental protocol do generate complex data: Eg. Tandem affinity purification (TAP) One may want to convert these complexes into sets of binary interactions, 2 algorithms are available:
How to deal with Complexes • • 13 Some experimental protocol do generate complex data: Eg. Tandem affinity purification (TAP) One may want to convert these complexes into sets of binary interactions, 2 algorithms are available:
 PSI-MI XML format • Community standard for Molecular Interactions • XML schema and detailed controlled vocabularies • Jointly developed by major data providers: BIND, Cell. Zome, DIP, GSK, HPRD, Hybrigenics, Int. Act, MINT, MIPS, Serono, U. Bielefeld, U. Bordeaux, U. Cambridge, and others • Version 1. 0 published in February 2004 The HUPO PSI Molecular Interaction Format - A community standard for the representation of protein interaction data. Henning Hermjakob et al, Nature Biotechnology 2004. • Version 2. 5 published in October 2007 Broadening the horizon - Level 2. 5 of the HUPO-PSI format for molecular interactions. Samuel Kerrien et al. , BMC Biology 2007.
PSI-MI XML format • Community standard for Molecular Interactions • XML schema and detailed controlled vocabularies • Jointly developed by major data providers: BIND, Cell. Zome, DIP, GSK, HPRD, Hybrigenics, Int. Act, MINT, MIPS, Serono, U. Bielefeld, U. Bordeaux, U. Cambridge, and others • Version 1. 0 published in February 2004 The HUPO PSI Molecular Interaction Format - A community standard for the representation of protein interaction data. Henning Hermjakob et al, Nature Biotechnology 2004. • Version 2. 5 published in October 2007 Broadening the horizon - Level 2. 5 of the HUPO-PSI format for molecular interactions. Samuel Kerrien et al. , BMC Biology 2007.
 Data distribution: PSICQUIC • Proteomics Standards Initiative Common QUery Interfa. Ce. • Community effort to standardise the way to access and retrieve data from Molecular Interaction databases. • Widely implemented by independent interaction data resources. • Based on the PSI standard formats (PSI-MI XML and MITAB) • Not limited to protein-protein interactions, also e. g. • Drug-target interactions • Simplified pathway data • A registry listing resources implementing PSICQUIC • Documentation: http: //psicquic. googlecode. com
Data distribution: PSICQUIC • Proteomics Standards Initiative Common QUery Interfa. Ce. • Community effort to standardise the way to access and retrieve data from Molecular Interaction databases. • Widely implemented by independent interaction data resources. • Based on the PSI standard formats (PSI-MI XML and MITAB) • Not limited to protein-protein interactions, also e. g. • Drug-target interactions • Simplified pathway data • A registry listing resources implementing PSICQUIC • Documentation: http: //psicquic. googlecode. com
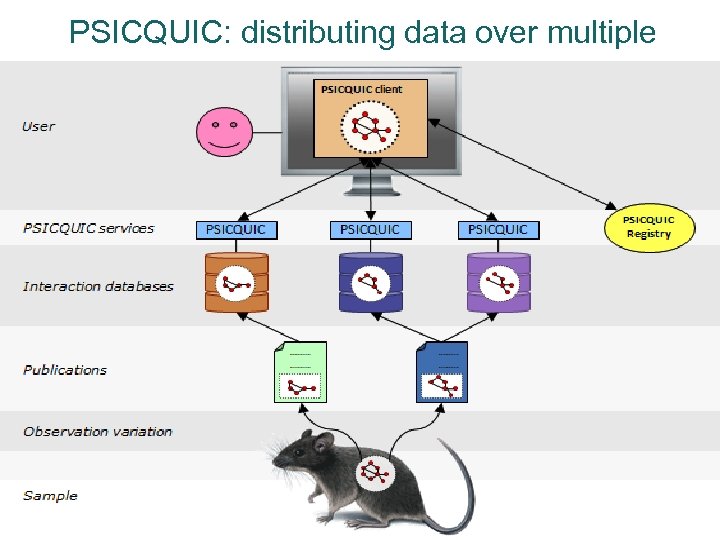 PSICQUIC: distributing data over multiple sources
PSICQUIC: distributing data over multiple sources
 IMEx: The International Molecular Exchange Consortium • Group of major public interaction data providers sharing curation effort: DIP, Int. Act, I 2 D, MINT, Matrix. DB, Molecular Connections, Innate. DB and MPIDB • Independent molecular interaction resources • Common curation standards for detailed curation • Common data formats (PSI-MI XML, PSICQUIC) • Common accession number space • Coordinated & non-redundant curation • In production mode since February 2010 • Since 3/2009 supported by the European Commission under PSIMEx, contract number FP 7 -HEALTH-2007 -223411, with additional partners Vital-IT, Nature, Wiley, Bia. Core (GE), U. Maryland, CSIC, TU Munich, MIPS, SCBIT (Shanghai) www. imexconsortium. org
IMEx: The International Molecular Exchange Consortium • Group of major public interaction data providers sharing curation effort: DIP, Int. Act, I 2 D, MINT, Matrix. DB, Molecular Connections, Innate. DB and MPIDB • Independent molecular interaction resources • Common curation standards for detailed curation • Common data formats (PSI-MI XML, PSICQUIC) • Common accession number space • Coordinated & non-redundant curation • In production mode since February 2010 • Since 3/2009 supported by the European Commission under PSIMEx, contract number FP 7 -HEALTH-2007 -223411, with additional partners Vital-IT, Nature, Wiley, Bia. Core (GE), U. Maryland, CSIC, TU Munich, MIPS, SCBIT (Shanghai) www. imexconsortium. org
 www. imexconsortium. org 18
www. imexconsortium. org 18
 Performing and visualing a Simple Search Data, Standards and Tools EBI Walthrough May 2009 EBI is an Outstation of the European Molecular Biology Laboratory.
Performing and visualing a Simple Search Data, Standards and Tools EBI Walthrough May 2009 EBI is an Outstation of the European Molecular Biology Laboratory.
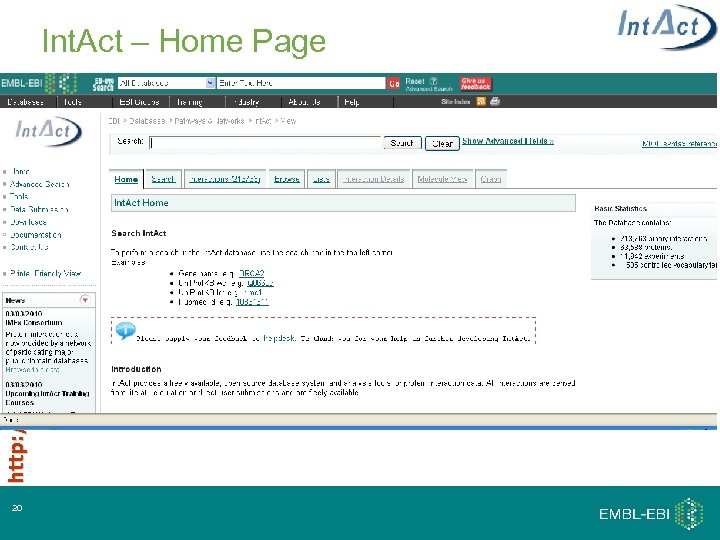 http: //www. ebi. ac. uk/intact Int. Act – Home Page 20
http: //www. ebi. ac. uk/intact Int. Act – Home Page 20
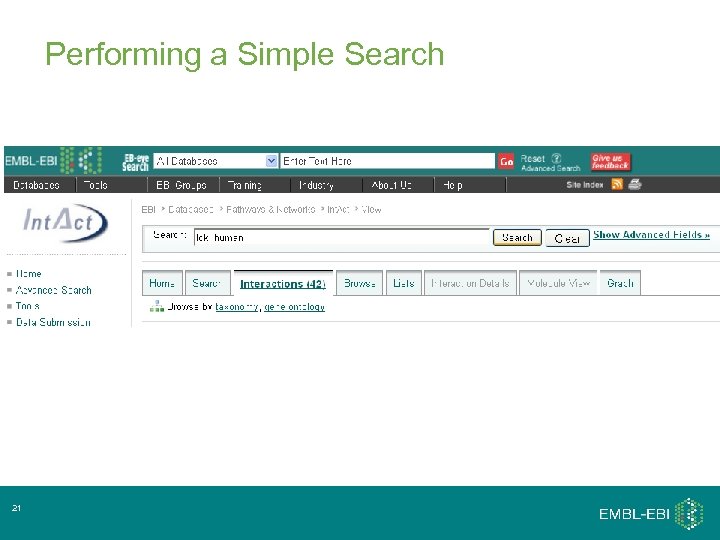 Performing a Simple Search 21
Performing a Simple Search 21
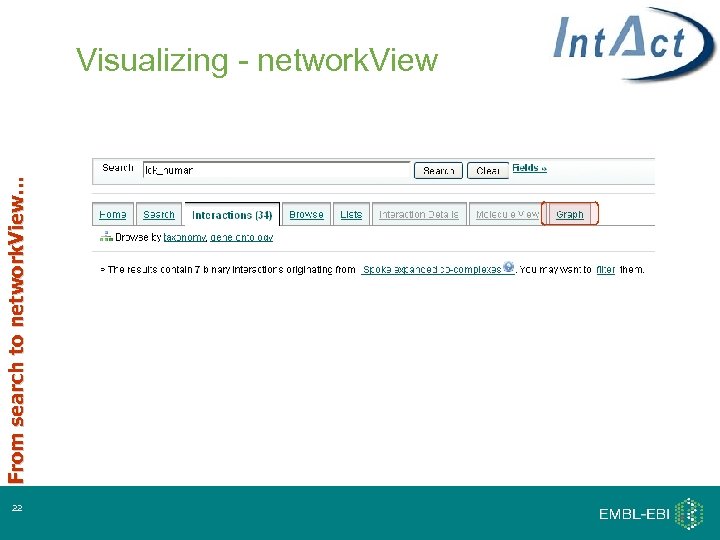 From search to network. View… Visualizing - network. View 22
From search to network. View… Visualizing - network. View 22
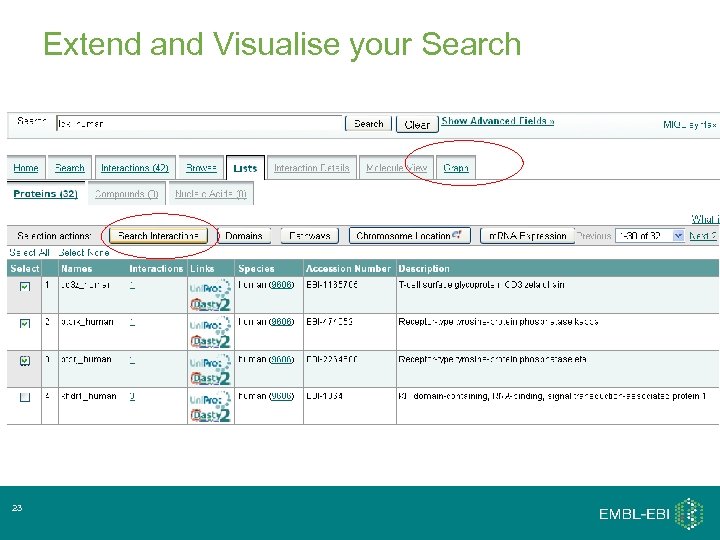 Extend and Visualise your Search 23
Extend and Visualise your Search 23
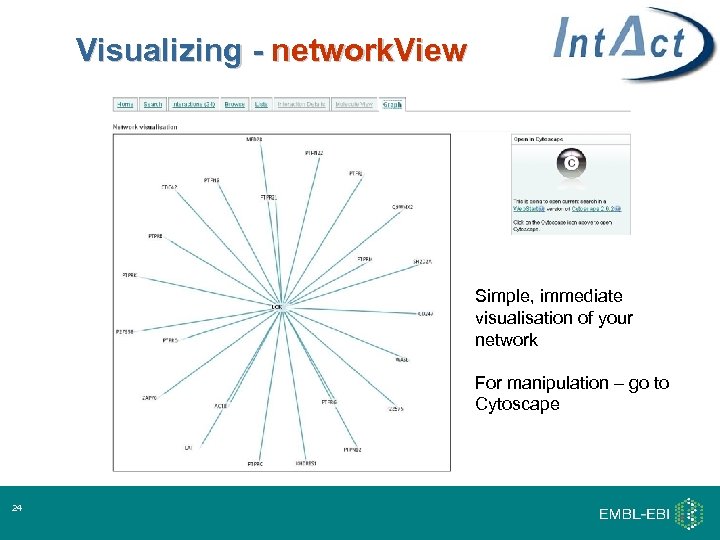 Visualizing - network. View Simple, immediate visualisation of your network For manipulation – go to Cytoscape 24
Visualizing - network. View Simple, immediate visualisation of your network For manipulation – go to Cytoscape 24
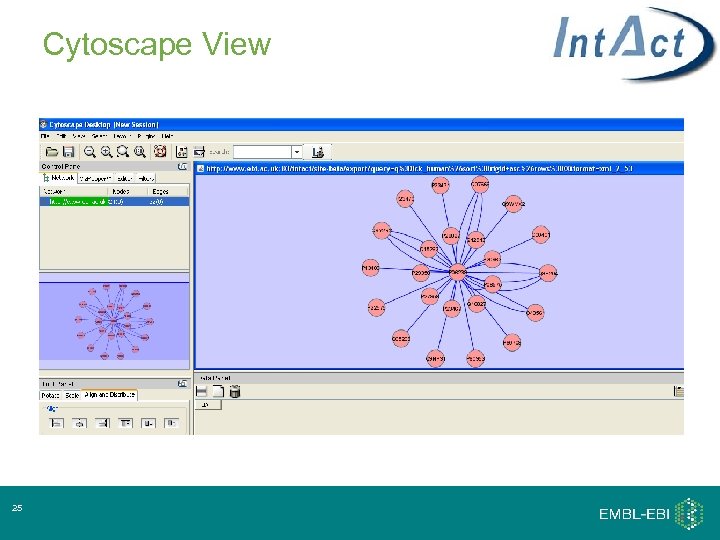 Cytoscape View 25
Cytoscape View 25
 Exploring a single interaction in more depth EBI is an Outstation of the European Molecular Biology Laboratory.
Exploring a single interaction in more depth EBI is an Outstation of the European Molecular Biology Laboratory.
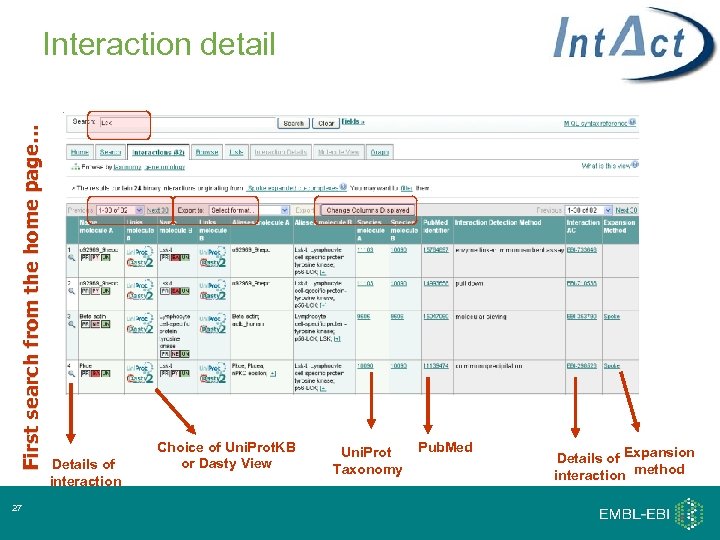 First search from the home page… Interaction detail 27 Details of interaction Choice of Uni. Prot. KB or Dasty View Uni. Prot Taxonomy Pub. Med Details of Expansion interaction method
First search from the home page… Interaction detail 27 Details of interaction Choice of Uni. Prot. KB or Dasty View Uni. Prot Taxonomy Pub. Med Details of Expansion interaction method
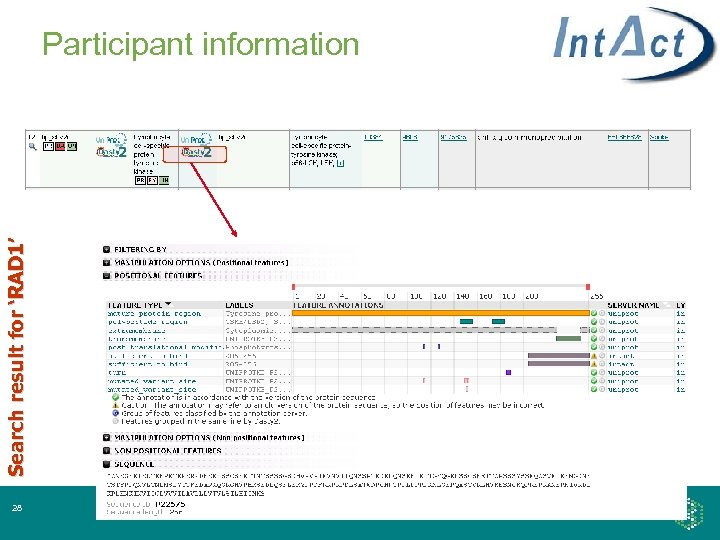 Search result for ‘RAD 1’ Participant information 28
Search result for ‘RAD 1’ Participant information 28
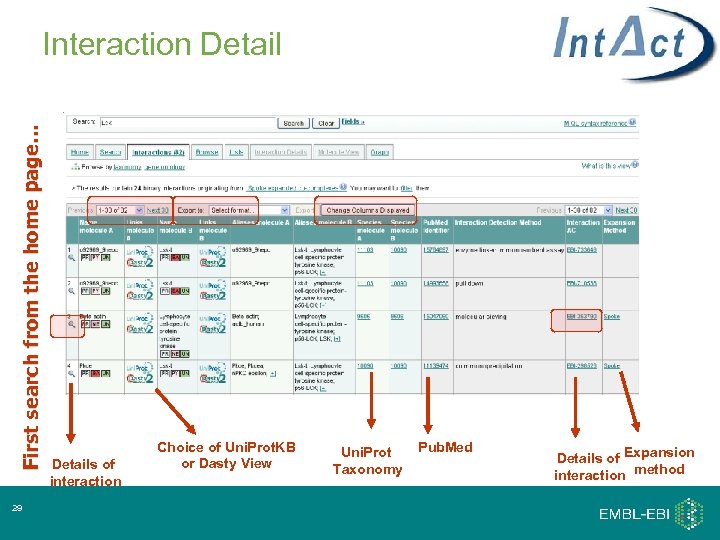 First search from the home page… Interaction Detail 29 Details of interaction Choice of Uni. Prot. KB or Dasty View Uni. Prot Taxonomy Pub. Med Details of Expansion interaction method
First search from the home page… Interaction Detail 29 Details of interaction Choice of Uni. Prot. KB or Dasty View Uni. Prot Taxonomy Pub. Med Details of Expansion interaction method
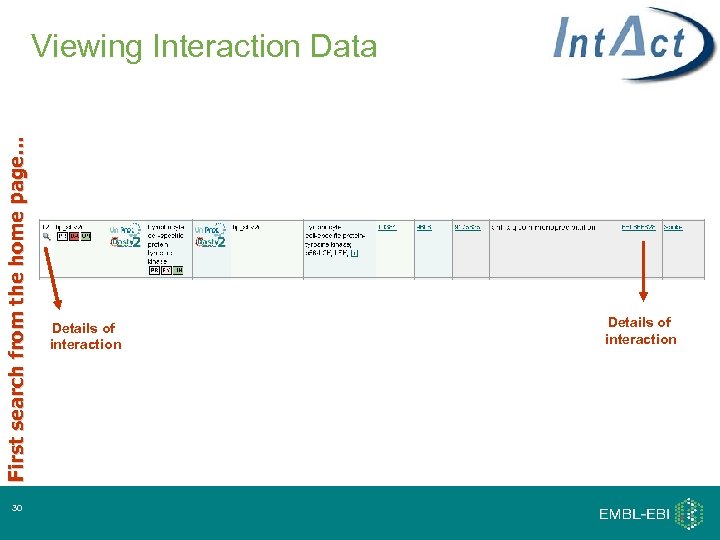 First search from the home page… Viewing Interaction Data 30 Details of interaction
First search from the home page… Viewing Interaction Data 30 Details of interaction
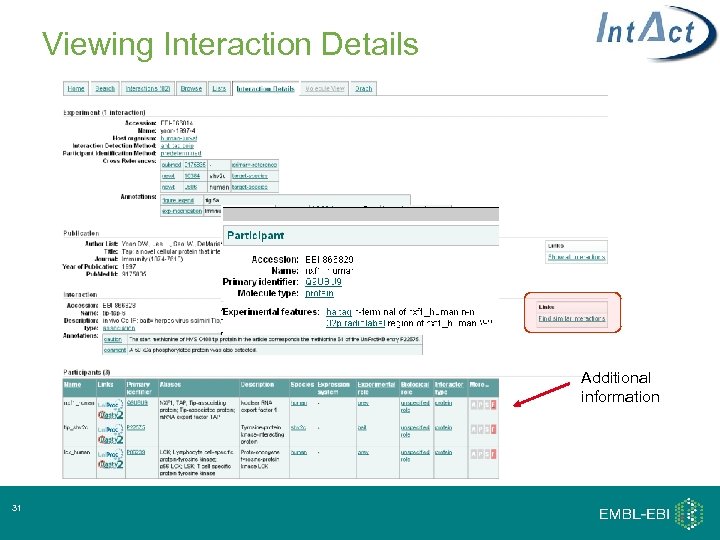 Viewing Interaction Details Additional information 31
Viewing Interaction Details Additional information 31
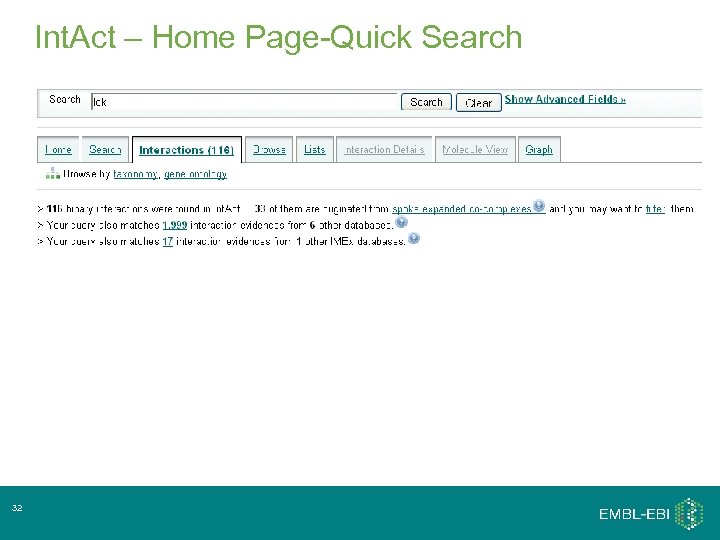 Int. Act – Home Page-Quick Search 32
Int. Act – Home Page-Quick Search 32
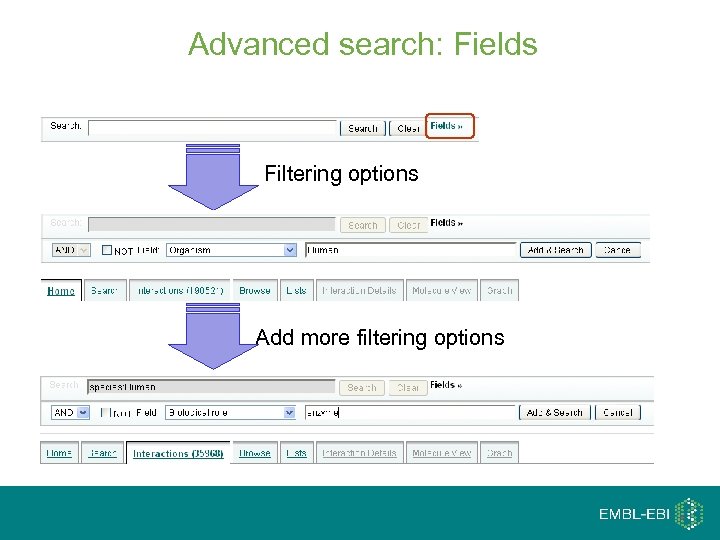 Advanced search: Fields Filtering options Add more filtering options
Advanced search: Fields Filtering options Add more filtering options
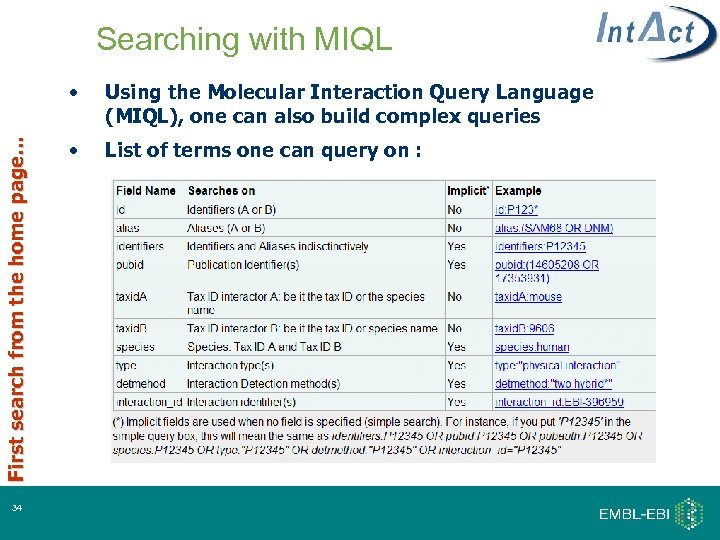 Searching with MIQL First search from the home page… • 34 Using the Molecular Interaction Query Language (MIQL), one can also build complex queries • List of terms one can query on :
Searching with MIQL First search from the home page… • 34 Using the Molecular Interaction Query Language (MIQL), one can also build complex queries • List of terms one can query on :
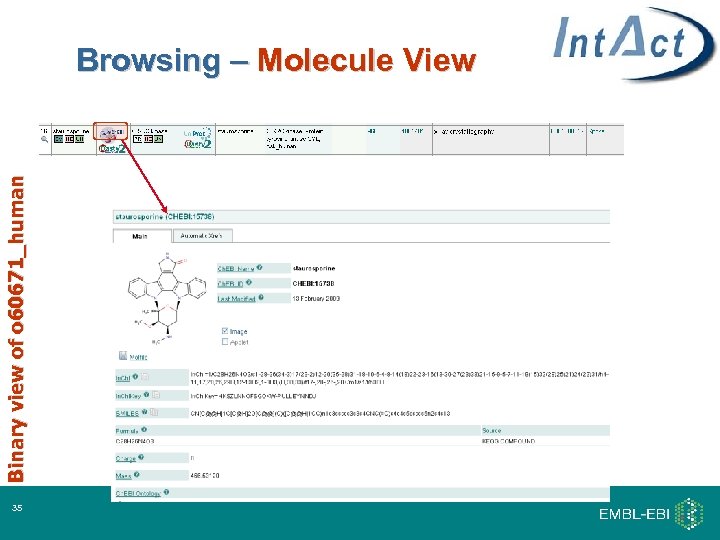 Binary view of o 60671_human Browsing – Molecule View 35
Binary view of o 60671_human Browsing – Molecule View 35
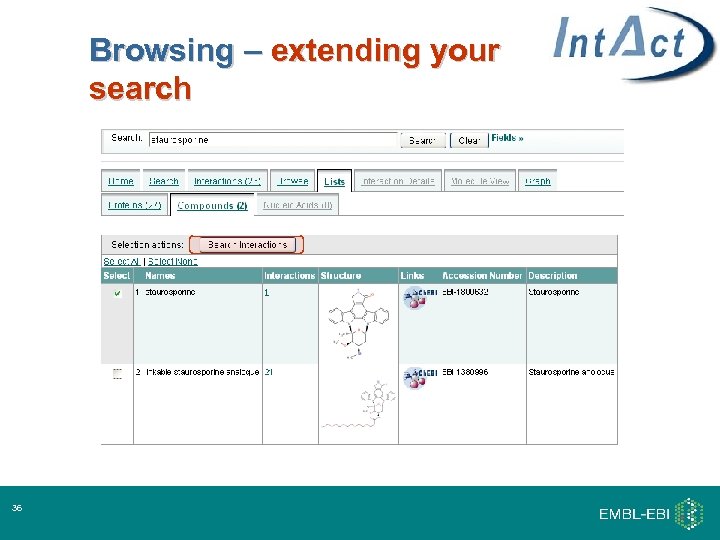 Browsing – extending your search 36
Browsing – extending your search 36
 ? ? ? ? ? ? 37 ? ? ?
? ? ? ? ? ? 37 ? ? ?
 An overview of the PSI-MI XML 2. 5 DATA MODEL 38
An overview of the PSI-MI XML 2. 5 DATA MODEL 38
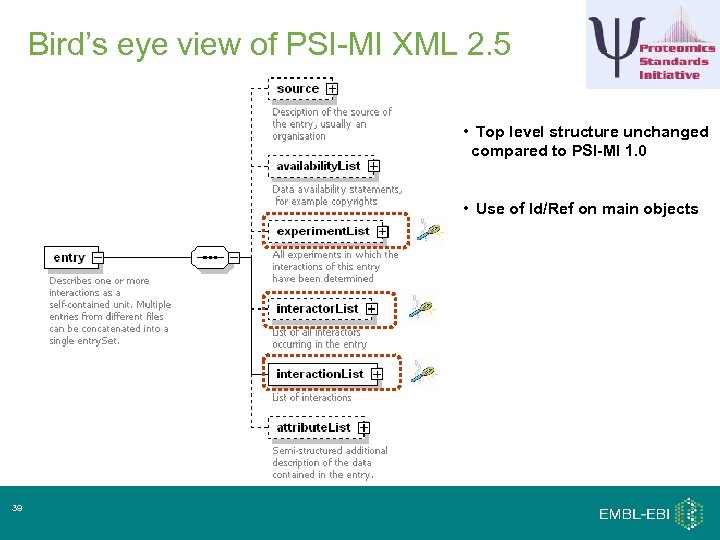 Bird’s eye view of PSI-MI XML 2. 5 • Top level structure unchanged compared to PSI-MI 1. 0 • Use of Id/Ref on main objects 39
Bird’s eye view of PSI-MI XML 2. 5 • Top level structure unchanged compared to PSI-MI 1. 0 • Use of Id/Ref on main objects 39
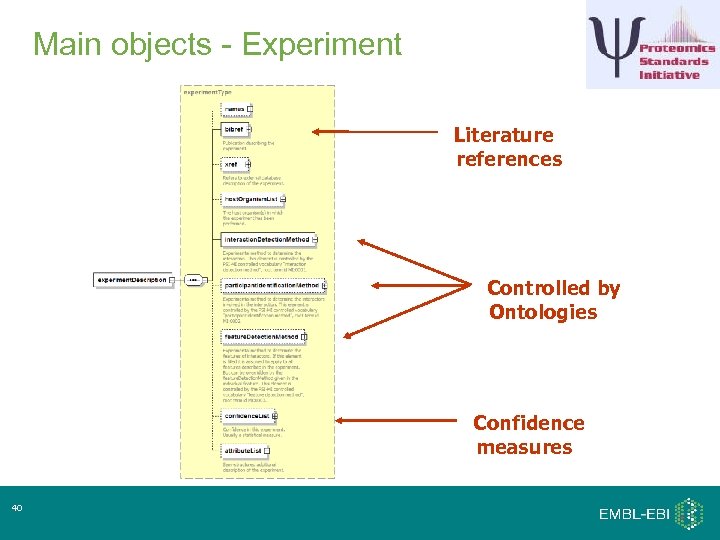 Main objects - Experiment Literature references Controlled by Ontologies Confidence measures 40
Main objects - Experiment Literature references Controlled by Ontologies Confidence measures 40
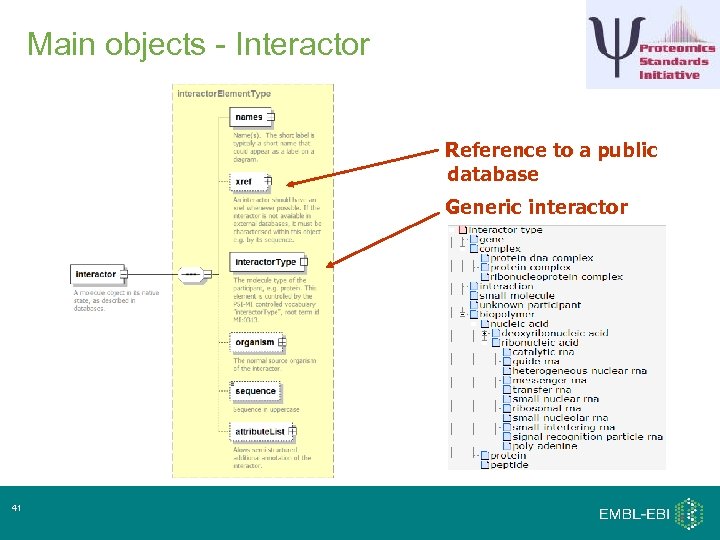 Main objects - Interactor Reference to a public database Generic interactor 41
Main objects - Interactor Reference to a public database Generic interactor 41
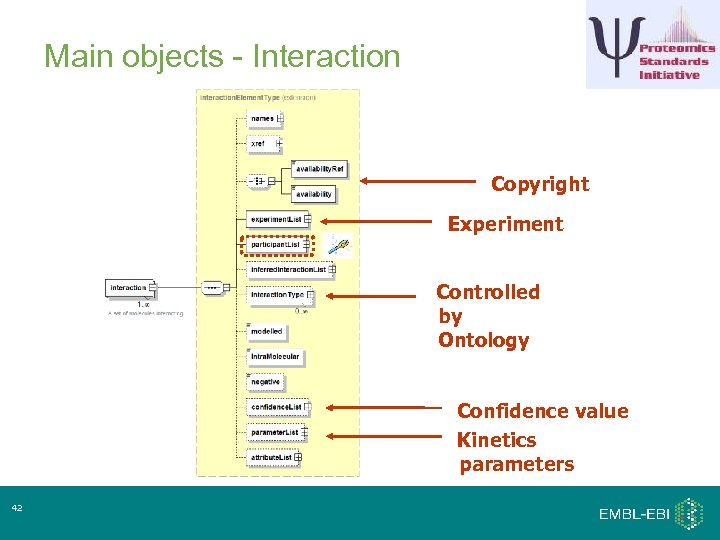 Main objects - Interaction Copyright Experiment Controlled by Ontology Confidence value Kinetics parameters 42
Main objects - Interaction Copyright Experiment Controlled by Ontology Confidence value Kinetics parameters 42
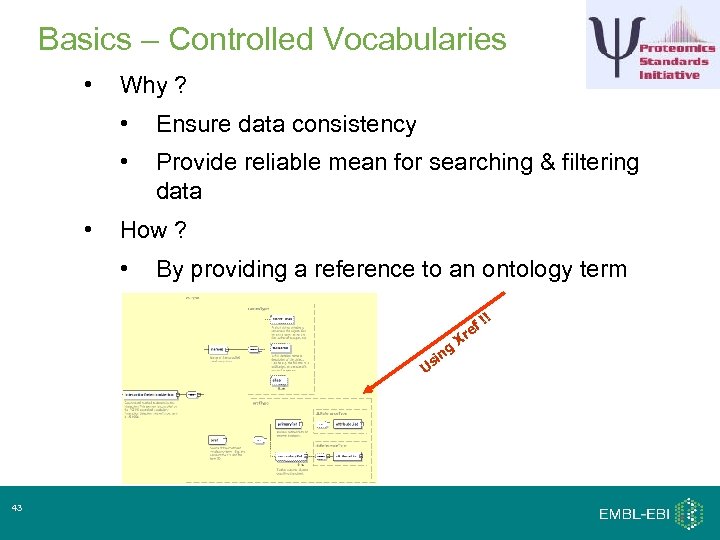 Basics – Controlled Vocabularies • Why ? • • • Ensure data consistency Provide reliable mean for searching & filtering data How ? • By providing a reference to an ontology term ! ! ef ng si U 43 Xr
Basics – Controlled Vocabularies • Why ? • • • Ensure data consistency Provide reliable mean for searching & filtering data How ? • By providing a reference to an ontology term ! ! ef ng si U 43 Xr
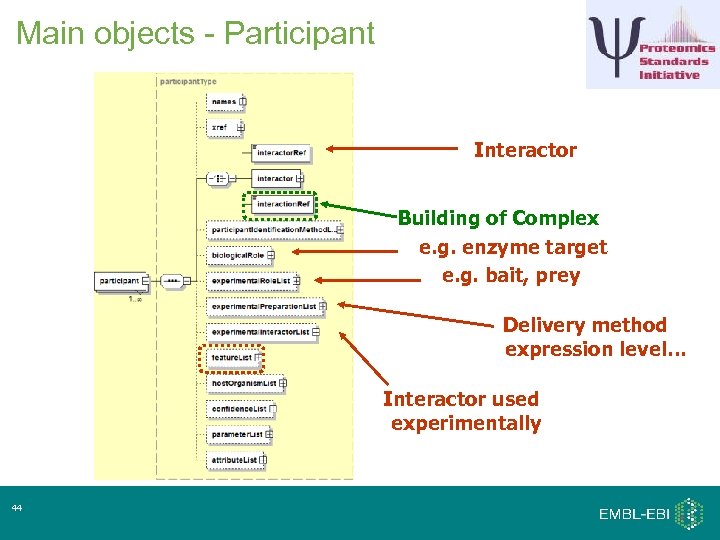 Main objects - Participant Interactor Building of Complex e. g. enzyme target e. g. bait, prey Delivery method expression level… Interactor used experimentally 44
Main objects - Participant Interactor Building of Complex e. g. enzyme target e. g. bait, prey Delivery method expression level… Interactor used experimentally 44
 An overview of the PSI-MI TAB DATA MODEL 45
An overview of the PSI-MI TAB DATA MODEL 45
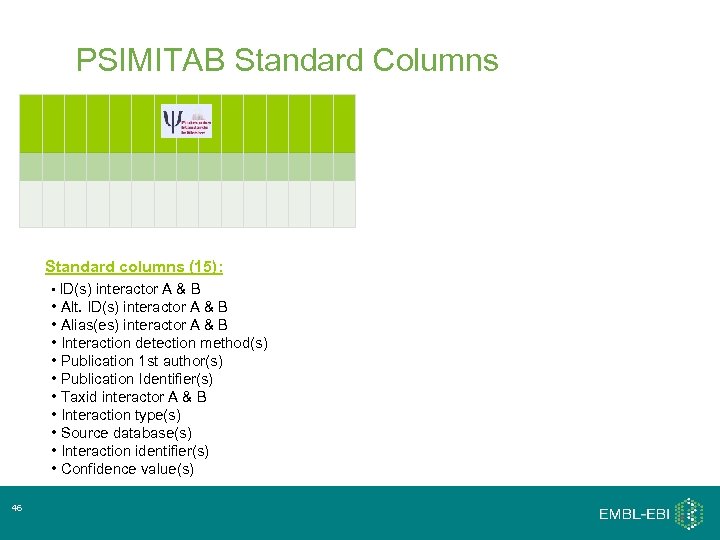 PSIMITAB Standard Columns Standard columns (15): • ID(s) interactor A & B • Alt. ID(s) interactor A & B • Alias(es) interactor A & B • Interaction detection method(s) • Publication 1 st author(s) • Publication Identifier(s) • Taxid interactor A & B • Interaction type(s) • Source database(s) • Interaction identifier(s) • Confidence value(s) 46
PSIMITAB Standard Columns Standard columns (15): • ID(s) interactor A & B • Alt. ID(s) interactor A & B • Alias(es) interactor A & B • Interaction detection method(s) • Publication 1 st author(s) • Publication Identifier(s) • Taxid interactor A & B • Interaction type(s) • Source database(s) • Interaction identifier(s) • Confidence value(s) 46
 A quick look into INTACT EXTENDED MITAB 47
A quick look into INTACT EXTENDED MITAB 47
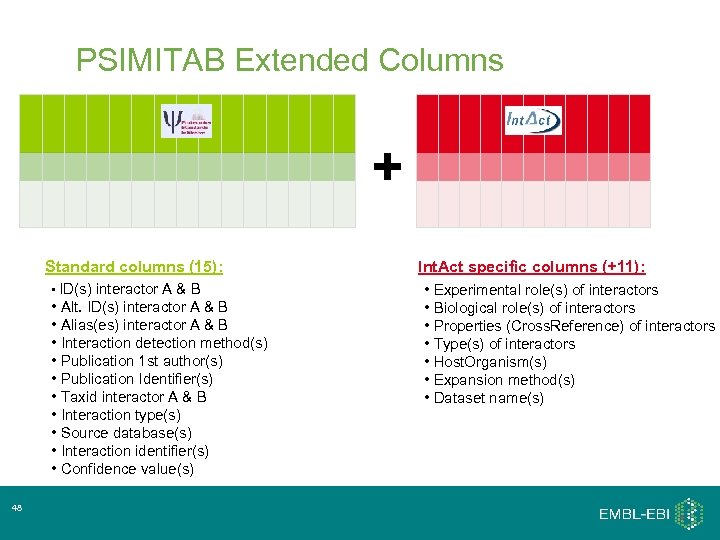 PSIMITAB Extended Columns + Standard columns (15): • ID(s) interactor A & B • Alt. ID(s) interactor A & B • Alias(es) interactor A & B • Interaction detection method(s) • Publication 1 st author(s) • Publication Identifier(s) • Taxid interactor A & B • Interaction type(s) • Source database(s) • Interaction identifier(s) • Confidence value(s) 48 Int. Act specific columns (+11): • Experimental role(s) of interactors • Biological role(s) of interactors • Properties (Cross. Reference) of interactors • Type(s) of interactors • Host. Organism(s) • Expansion method(s) • Dataset name(s)
PSIMITAB Extended Columns + Standard columns (15): • ID(s) interactor A & B • Alt. ID(s) interactor A & B • Alias(es) interactor A & B • Interaction detection method(s) • Publication 1 st author(s) • Publication Identifier(s) • Taxid interactor A & B • Interaction type(s) • Source database(s) • Interaction identifier(s) • Confidence value(s) 48 Int. Act specific columns (+11): • Experimental role(s) of interactors • Biological role(s) of interactors • Properties (Cross. Reference) of interactors • Type(s) of interactors • Host. Organism(s) • Expansion method(s) • Dataset name(s)
 A hands on introduction to PSI-MI XML 2. 5 JAVA API 49
A hands on introduction to PSI-MI XML 2. 5 JAVA API 49
 PSI-MI XML Java API • • Uses Java 5 Provides binding between XML and Java object model Tools to read/write XML from/to file Read can be done in 2 fashions: • Load a whole file in an Entry. Set • Only allows to load large files if you have enough memory • Easy to update content and write back to file • Index XML data and give access though an Indexed. Entry • Memory efficient with large files • Allows to browse through interactions, experiments… • Trickier to write updated content (yet, feasible) 50
PSI-MI XML Java API • • Uses Java 5 Provides binding between XML and Java object model Tools to read/write XML from/to file Read can be done in 2 fashions: • Load a whole file in an Entry. Set • Only allows to load large files if you have enough memory • Easy to update content and write back to file • Index XML data and give access though an Indexed. Entry • Memory efficient with large files • Allows to browse through interactions, experiments… • Trickier to write updated content (yet, feasible) 50
 A hands on introduction to PSI-MI TAB 2. 5 JAVA API 51
A hands on introduction to PSI-MI TAB 2. 5 JAVA API 51
 PSI-MI TAB Java API • • Uses Java 5 Provides binding between TAB and a Java object model Tools to read/write TAB from/to file You can read in 2 fashions: • Load a whole file in a Collection
PSI-MI TAB Java API • • Uses Java 5 Provides binding between TAB and a Java object model Tools to read/write TAB from/to file You can read in 2 fashions: • Load a whole file in a Collection
 Summary • PSI-MI XML is the de facto standard for molecular interactions • We have code samples & exercises for both APIs ! Let me know if you want access to it … • The Java API makes it easy to handle PSI-MI Home page http: //psidev. info/MI API Download http: //www. psidev. info/index. php? q=node/60#tools Data ftp: //ftp. ebi. ac. uk/pub/databases/intact/current/psi 25 53
Summary • PSI-MI XML is the de facto standard for molecular interactions • We have code samples & exercises for both APIs ! Let me know if you want access to it … • The Java API makes it easy to handle PSI-MI Home page http: //psidev. info/MI API Download http: //www. psidev. info/index. php? q=node/60#tools Data ftp: //ftp. ebi. ac. uk/pub/databases/intact/current/psi 25 53
 Quick introduction to R packages for PSI-MI 54
Quick introduction to R packages for PSI-MI 54
 Rintact & Rpsi. XML • Initiative from the Wolfgang Huber’s group at the EBI • Enables PSI-MI XML data read into R data structure • Enables data analysis using existing packages such as: RBGL, ppi. Stats, ap. Complex, … • Currently supports: Int. Act, MINT, HPRD, DIP, Bio. GRID, MIPS/CORUM, Matri. DB, MPACT. API Download http: //www. bioconductor. org/packages/2. 1/bioc/html/Rintact. html Documentation http: //www. bioconductor. org/packages/2. 3/bioc/vignettes/Rpsi. XML/inst/doc/Rpsi. XML. pdf 55
Rintact & Rpsi. XML • Initiative from the Wolfgang Huber’s group at the EBI • Enables PSI-MI XML data read into R data structure • Enables data analysis using existing packages such as: RBGL, ppi. Stats, ap. Complex, … • Currently supports: Int. Act, MINT, HPRD, DIP, Bio. GRID, MIPS/CORUM, Matri. DB, MPACT. API Download http: //www. bioconductor. org/packages/2. 1/bioc/html/Rintact. html Documentation http: //www. bioconductor. org/packages/2. 3/bioc/vignettes/Rpsi. XML/inst/doc/Rpsi. XML. pdf 55


