2859d3cdc2adf736595518d302f4a70c.ppt
- Количество слайдов: 150

25| DNA Metabolism © 2017 W. H. Freeman and Company

CHAPTER 25 DNA Metabolism Learning goals: • DNA replication • DNA repair • DNA recombination
What Is DNA Metabolism? • Although DNA provides stable storage of genetic information, the structure is far from static: – A new copy of DNA is synthesized with high fidelity before each cell division. – Errors that arise during or after DNA synthesis are constantly checked for, and repairs are made. – Segments of DNA are rearranged either within a chromosome or between two DNA molecules (recombination), giving offspring a novel DNA. • DNA metabolism consists of a set of tightly regulated processes that achieve these tasks.
Bacterial Gene Naming • Three italicized lowercase letters • Example: uvr • Name usually reflects function – uvr encodes gene for resistance to UV radiation • Capital letters added to abbreviation reflect order of discovery, not enzymatic order – dna. A, dna. B genes for replication
Bacterial Protein Naming • • Often named after their genes Nonitalicized, roman type First letter capitalized Example: Dna. A is the protein encoded by the gene dna. A.
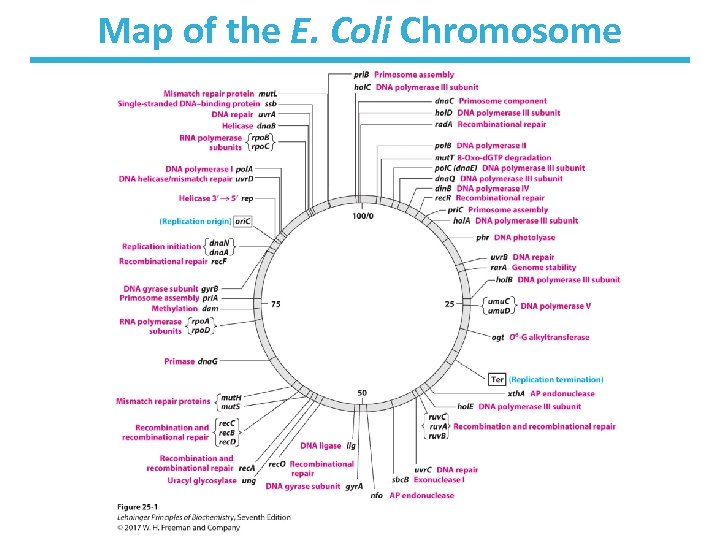
Map of the E. Coli Chromosome

Eukaryotic Gene Naming • • Not universal for all eukaryotic organisms Can vary with each species Name usually reflects function In Saccharomyces cerevisiae, gene names typically three uppercase italicized letters followed by a number – COX 1 – gene that codes for a subunit of cytochrome oxidase
Eukaryotic Protein Naming • Complex and varies for each species • May have the same name as the gene but the case of the letters is different – In S. cerevisiae, the protein has the first letter capitalized followed by two lower case letters, the number, and the letter “p. ” – Example: RAD 51 (gene) – Rad 51 p (protein)
DNA Replication Properties • Three fundamental rules of replication – Replication is semiconservative – Replication begins at an origin and proceeds (usually) bidirectionally – Synthesis of new DNA occurs in the 5’ 3’ direction and is semidiscontinuous
DNA Replication Is Semiconservative • The Meselson-Stahl experiment (next slide) showed that the nitrogen used for the synthesis of new ds. DNA becomes equally divided between the two daughter genomes. • This result suggested a semiconservative replication mechanism. – Each new DNA has one old (parent) strand one new (daughter) strand.
The Meselson-Stahl Experiment • Proved the hypothesis of semiconservative replication proposed by Watson and Crick • Cells were grown on a medium containing only 15 N isotope (heavy N) until fully labeled. – produced ONE band when DNA is centrifuged in Cs. Cl • Cells were then switched to 14 N medium and allowed to divide once. – ONE band but at a higher position than 15 N DNA, but lower than completely 14 N DNA ( hybrid DNA) • Cells were allowed to divide once more. – TWO bands, one with all 14 N DNA, one hybrid
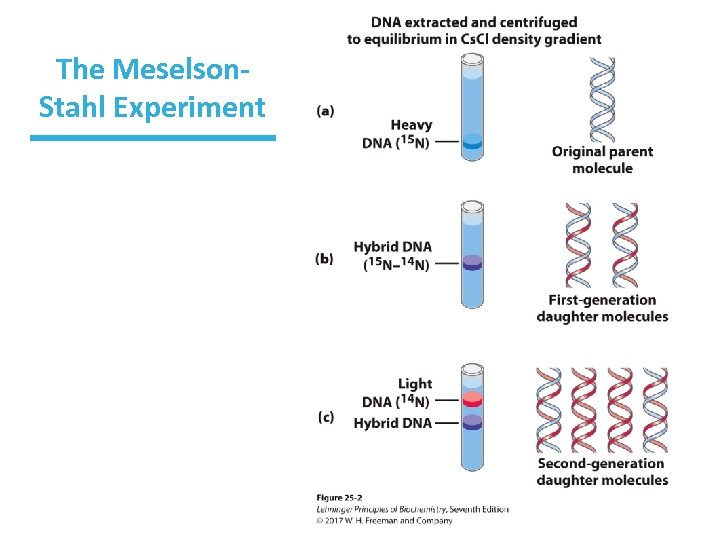
The Meselson. Stahl Experiment

How Did This Experiment Prove Semiconservative Replication? • Alternative hypothesis: conservative replication (two daughter strands would be completely new DNA) There would have been no intermediate hybrid band if this hypothesis were true.
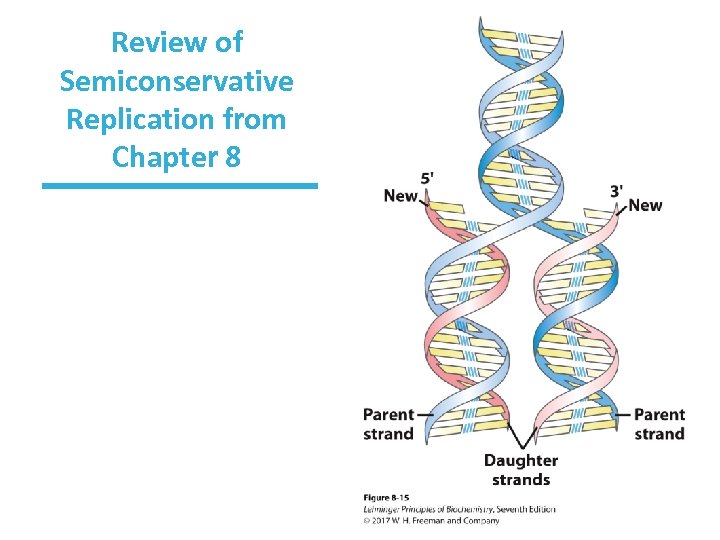
Review of Semiconservative Replication from Chapter 8
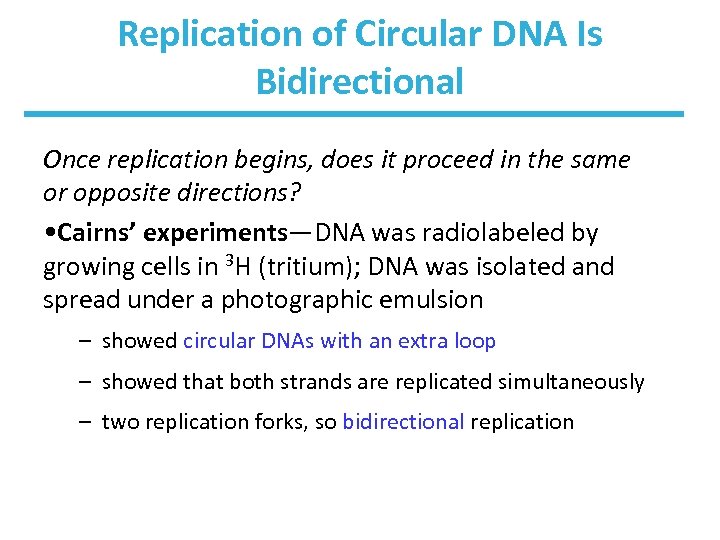
Replication of Circular DNA Is Bidirectional Once replication begins, does it proceed in the same or opposite directions? • Cairns’ experiments―DNA was radiolabeled by growing cells in 3 H (tritium); DNA was isolated and spread under a photographic emulsion – showed circular DNAs with an extra loop – showed that both strands are replicated simultaneously – two replication forks, so bidirectional replication
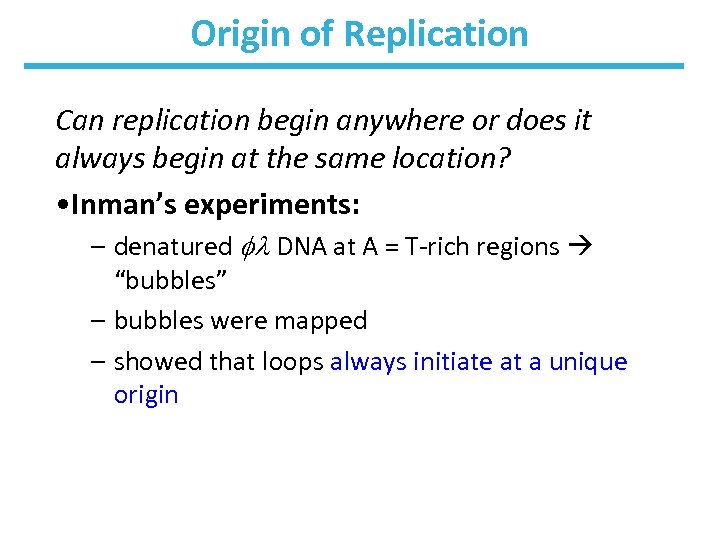
Origin of Replication Can replication begin anywhere or does it always begin at the same location? • Inman’s experiments: – denatured DNA at A = T-rich regions “bubbles” – bubbles were mapped – showed that loops always initiate at a unique origin
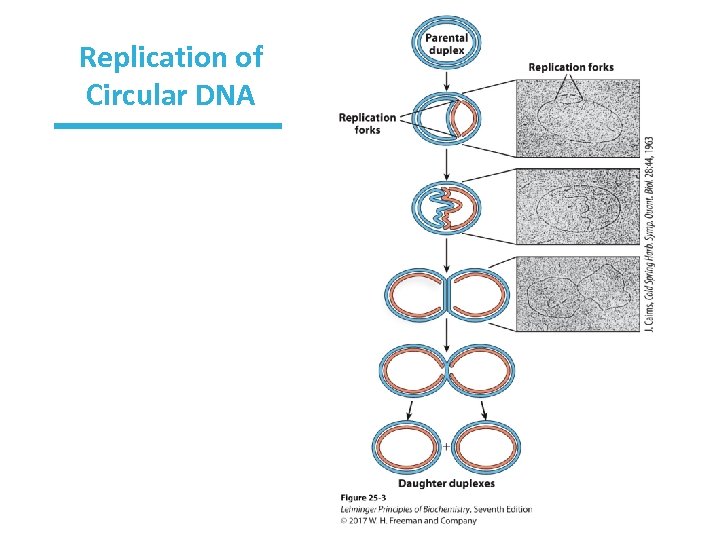
Replication of Circular DNA

Synthesis Proceeds in the Direction 5’ 3’ • Synthesis always occurs by addition of new nucleotides to the 3’ end (3’-OH). • The leading strand is made continuously as the replication fork advances. • The lagging strand is made discontinuously in short pieces (Okazaki fragments) that are later joined together.
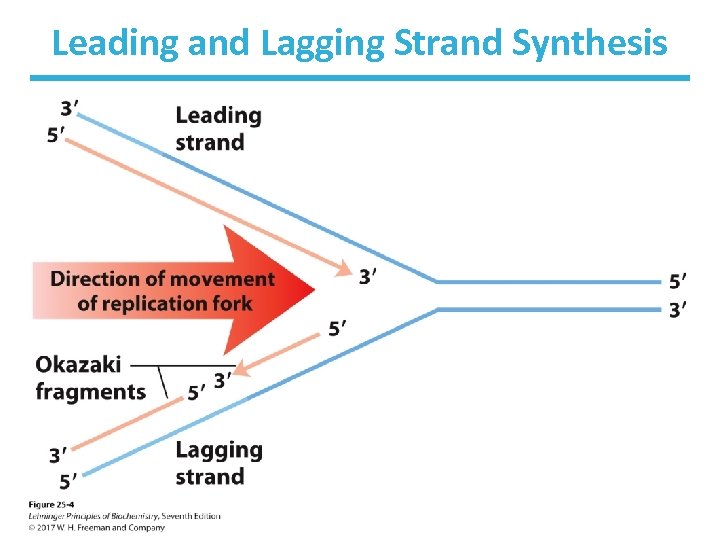
Leading and Lagging Strand Synthesis
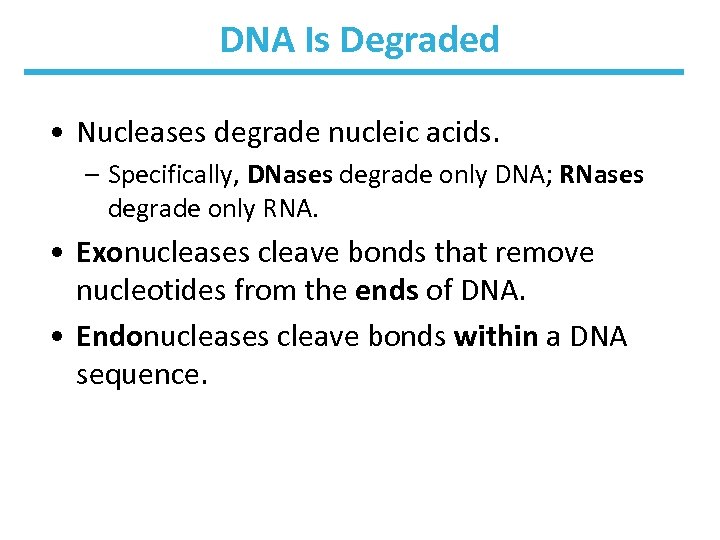
DNA Is Degraded • Nucleases degrade nucleic acids. – Specifically, DNases degrade only DNA; RNases degrade only RNA. • Exonucleases cleave bonds that remove nucleotides from the ends of DNA. • Endonucleases cleave bonds within a DNA sequence.
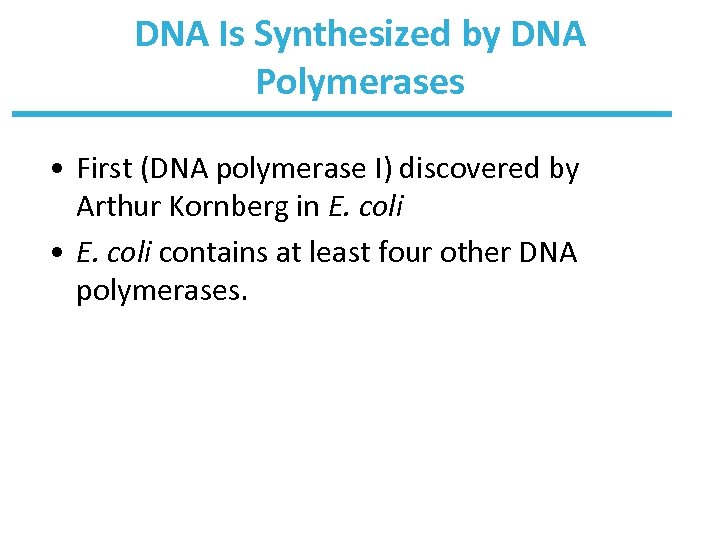
DNA Is Synthesized by DNA Polymerases • First (DNA polymerase I) discovered by Arthur Kornberg in E. coli • E. coli contains at least four other DNA polymerases.
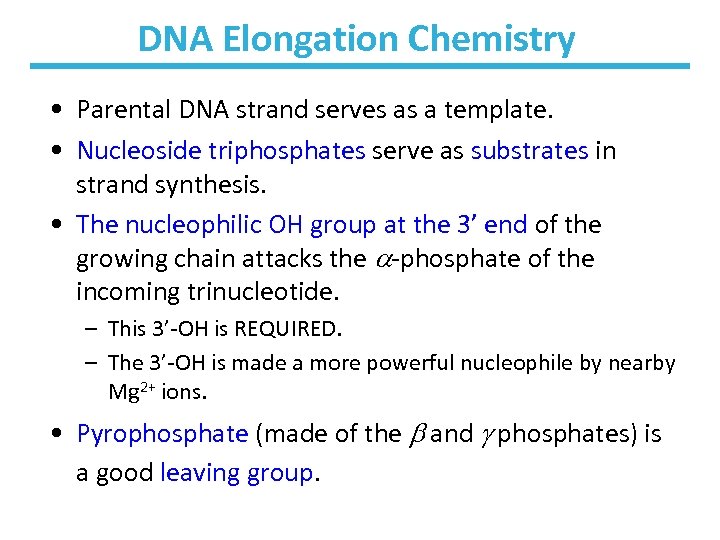
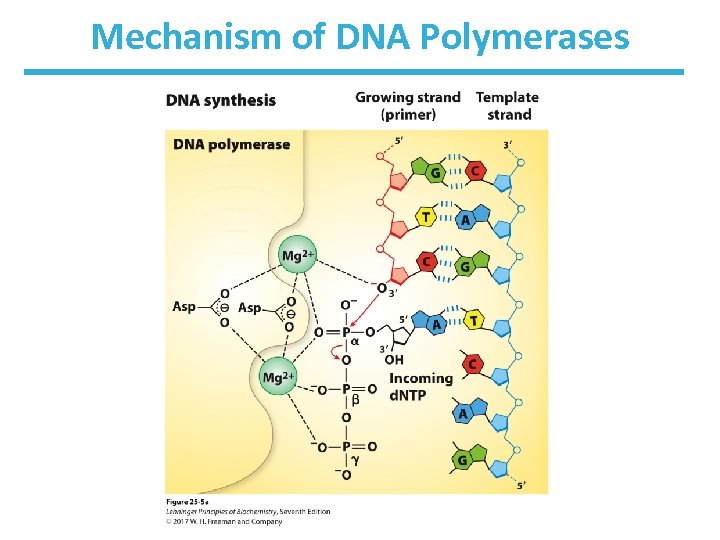
DNA Elongation Chemistry • Parental DNA strand serves as a template. • Nucleoside triphosphates serve as substrates in strand synthesis. • The nucleophilic OH group at the 3’ end of the growing chain attacks the -phosphate of the incoming trinucleotide. – This 3’-OH is REQUIRED. – The 3’-OH is made a more powerful nucleophile by nearby Mg 2+ ions. • Pyrophosphate (made of the and phosphates) is a good leaving group.
Mechanism of DNA Polymerases
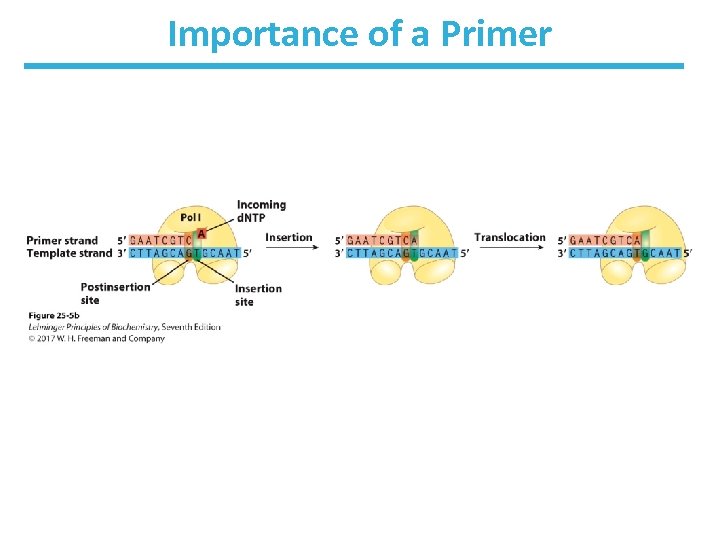
DNA Polymerase Also Requires a Primer • Primer = short strand complementary to the template – contains a 3’-OH to begin the first DNA polymerase-catalyzed reaction – can be made of DNA or RNA (more common)
Importance of a Primer
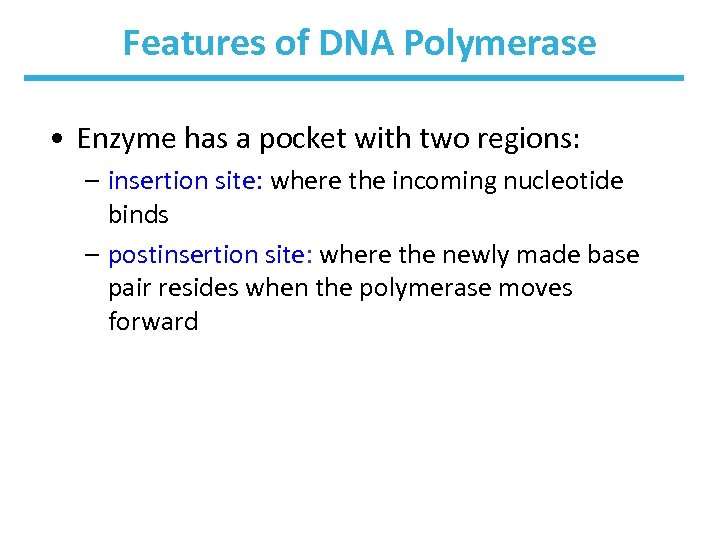
Features of DNA Polymerase • Enzyme has a pocket with two regions: – insertion site: where the incoming nucleotide binds – postinsertion site: where the newly made base pair resides when the polymerase moves forward
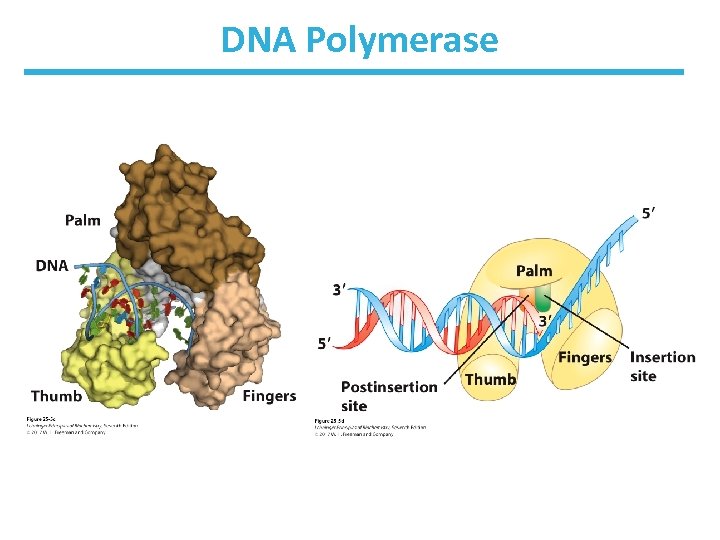
DNA Polymerase
DNA Polymerase Can Add Nucleotides or Dissociate • The number of nucleotides added before dissociation is called processivity. • The processivity of polymerases, in general, varies widely from a few nucleotides to many thousands. • Each specific polymerase has its own processivity and polymerization rate.
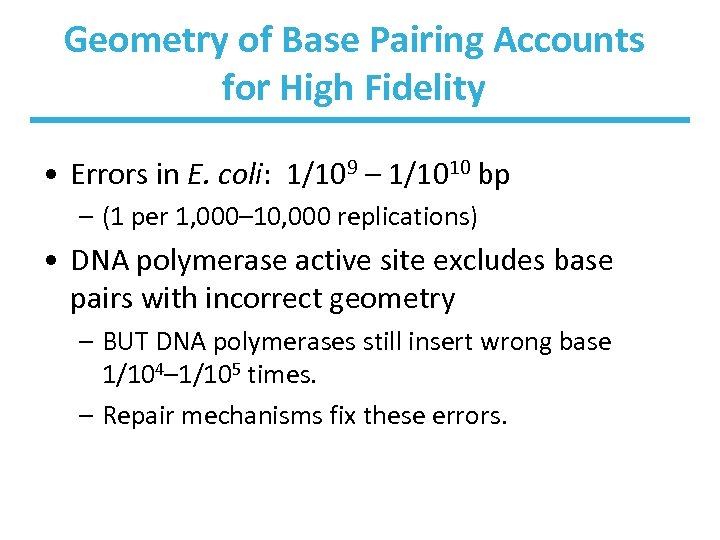
Geometry of Base Pairing Accounts for High Fidelity • Errors in E. coli: 1/109 – 1/1010 bp – (1 per 1, 000– 10, 000 replications) • DNA polymerase active site excludes base pairs with incorrect geometry – BUT DNA polymerases still insert wrong base 1/104– 1/105 times. – Repair mechanisms fix these errors.
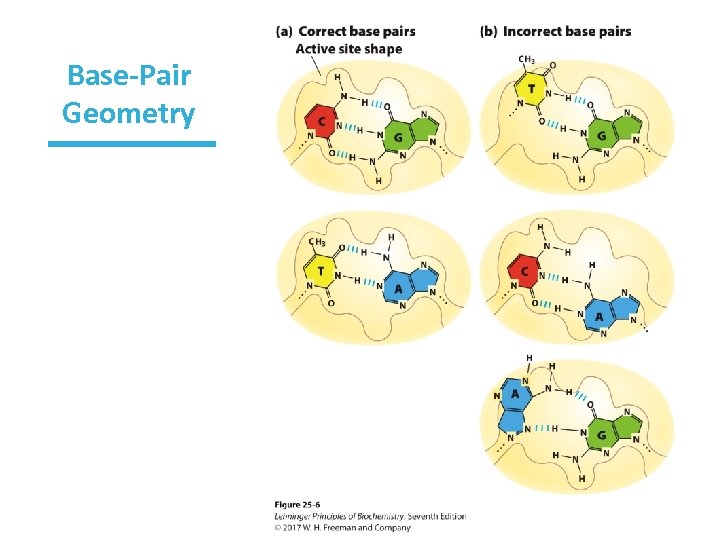
Base-Pair Geometry
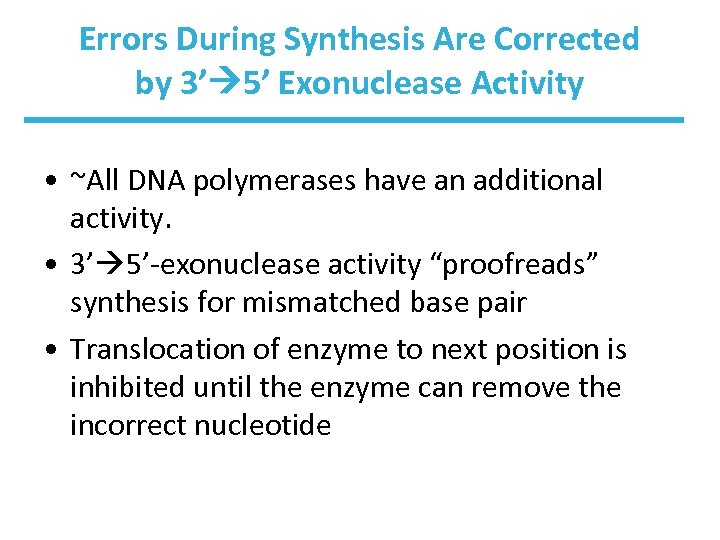
Errors During Synthesis Are Corrected by 3’ 5’ Exonuclease Activity • ~All DNA polymerases have an additional activity. • 3’ 5’-exonuclease activity “proofreads” synthesis for mismatched base pair • Translocation of enzyme to next position is inhibited until the enzyme can remove the incorrect nucleotide
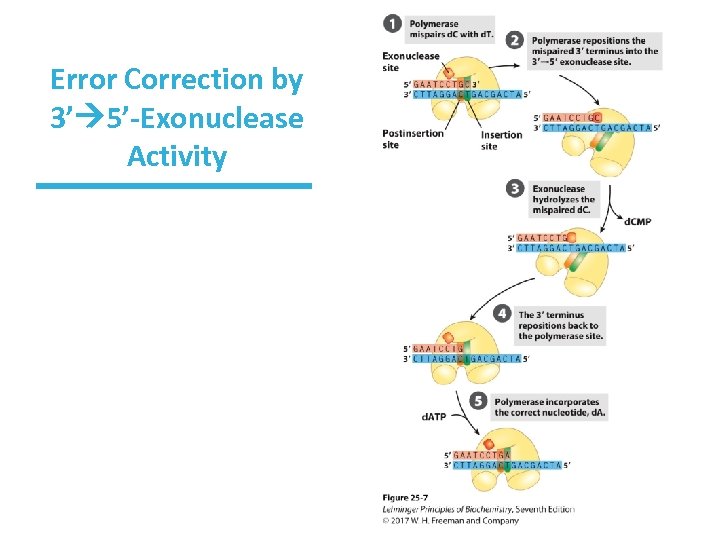
Error Correction by 3’ 5’-Exonuclease Activity
There Are at Least Five DNA Polymerases in E. Coli • DNA polymerase I is abundant but is not ideal for replication. – rate (600 nucleotides/min) is slower than observed for replication fork movement – has low processivity – primary function is in clean-up • DNA polymerase III is the principal replication polymerase. • DNA polymerases II, IV, and V are involved in DNA repair.
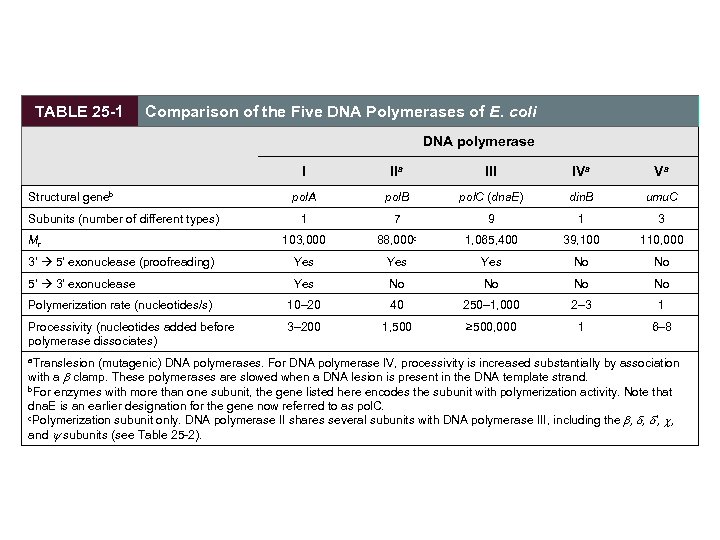
TABLE 25 -1 Comparison of the Five DNA Polymerases of E. coli DNA polymerase I IIa III IVa Va pol. A pol. B pol. C (dna. E) din. B umu. C 1 7 9 1 3 103, 000 88, 000 c 1, 065, 400 39, 100 110, 000 3' 5' exonuclease (proofreading) Yes Yes No No 5' 3' exonuclease Yes No No Polymerization rate (nucleotides/s) 10– 20 40 250– 1, 000 2– 3 1 Processivity (nucleotides added before polymerase dissociates) 3– 200 1, 500 ≥ 500, 000 1 6– 8 Structural geneb Subunits (number of different types) Mr a. Translesion (mutagenic) DNA polymerases. For DNA polymerase IV, processivity is increased substantially by association with a clamp. These polymerases are slowed when a DNA lesion is present in the DNA template strand. b. For enzymes with more than one subunit, the gene listed here encodes the subunit with polymerization activity. Note that dna. E is an earlier designation for the gene now referred to as pol. C. c. Polymerization subunit only. DNA polymerase II shares several subunits with DNA polymerase III, including the , , ', c, and y subunits (see Table 25 -2).
DNA Polymerase I Also Has 5’ 3’Exonuclease Activity • In addition to the 3’ 5’-exonuclease activity • Moves ahead of the enzyme, hydrolyzes things in its path • Does nick translation―a strand break moves along with enzyme • This activity and the polymerase activity are in the Klenow fragment―a distinct domain that can be separated by protease treatment.
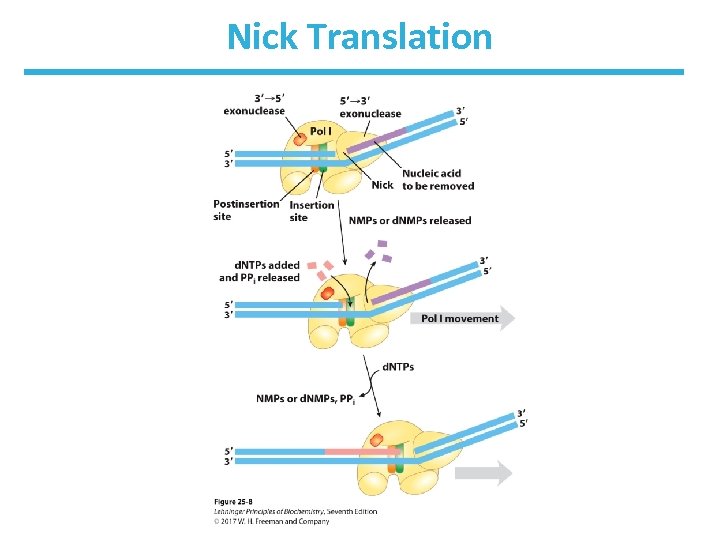
Nick Translation
DNA Polymerase III • Complex structure with 10 types of subunits • Two core domains of , , and subunits • The core domains are linked by the “clamploader” complex 2 ’. • The core domains each interact with a dimer of subunits that increase the processivity of the complex. – form a sliding clamp the prevents dissociation – processivity of DNA Pol III is >500, 000 bp because of the clamps
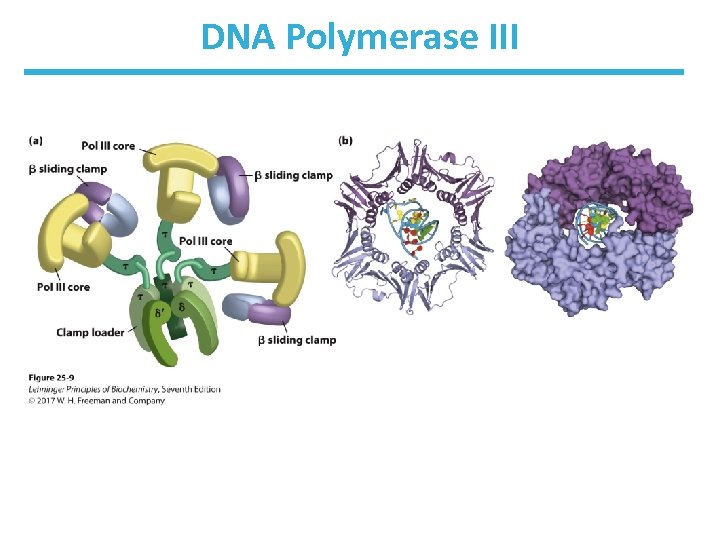
DNA Polymerase III
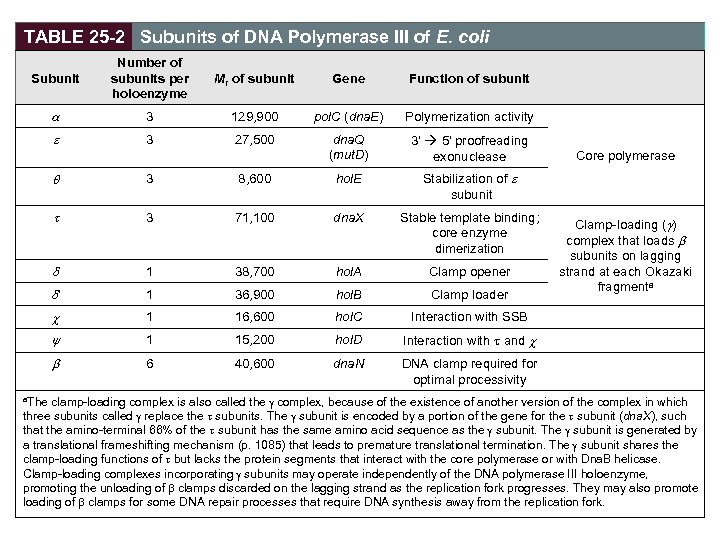
TABLE 25 -2 Subunits of DNA Polymerase III of E. coli Subunit Number of subunits per holoenzyme Mr of subunit Gene Function of subunit 3 129, 900 pol. C (dna. E) Polymerization activity 3 27, 500 dna. Q (mut. D) 3' 5' proofreading exonuclease 3 8, 600 hol. E Stabilization of subunit 3 71, 100 dna. X Stable template binding; core enzyme dimerization 1 38, 700 hol. A Clamp opener ' 1 36, 900 hol. B Clamp loader c 1 16, 600 hol. C Interaction with SSB y 1 15, 200 hol. D Interaction with t and c 6 40, 600 dna. N Core polymerase DNA clamp required for optimal processivity Clamp-loading ( ) complex that loads subunits on lagging strand at each Okazaki fragmenta clamp-loading complex is also called the g complex, because of the existence of another version of the complex in which three subunits called g replace the t subunits. The g subunit is encoded by a portion of the gene for the t subunit (dna. X), such that the amino-terminal 66% of the t subunit has the same amino acid sequence as the g subunit. The g subunit is generated by a translational frameshifting mechanism (p. 1085) that leads to premature translational termination. The g subunit shares the clamp-loading functions of t but lacks the protein segments that interact with the core polymerase or with Dna. B helicase. Clamp-loading complexes incorporating g subunits may operate independently of the DNA polymerase III holoenzyme, promoting the unloading of b clamps discarded on the lagging strand as the replication fork progresses. They may also promote loading of b clamps for some DNA repair processes that require DNA synthesis away from the replication fork. a. The
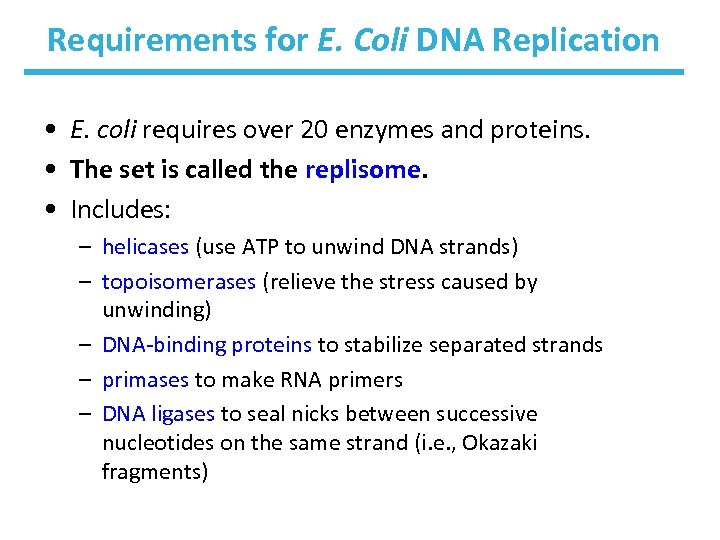
Requirements for E. Coli DNA Replication • E. coli requires over 20 enzymes and proteins. • The set is called the replisome. • Includes: – helicases (use ATP to unwind DNA strands) – topoisomerases (relieve the stress caused by unwinding) – DNA-binding proteins to stabilize separated strands – primases to make RNA primers – DNA ligases to seal nicks between successive nucleotides on the same strand (i. e. , Okazaki fragments)
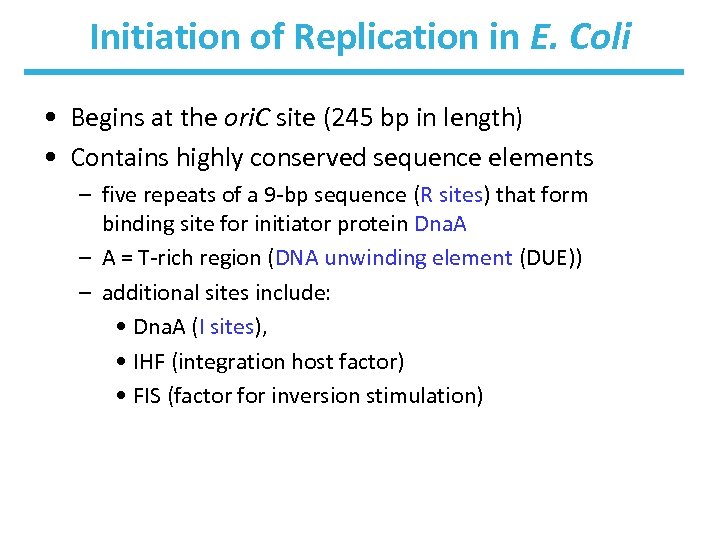
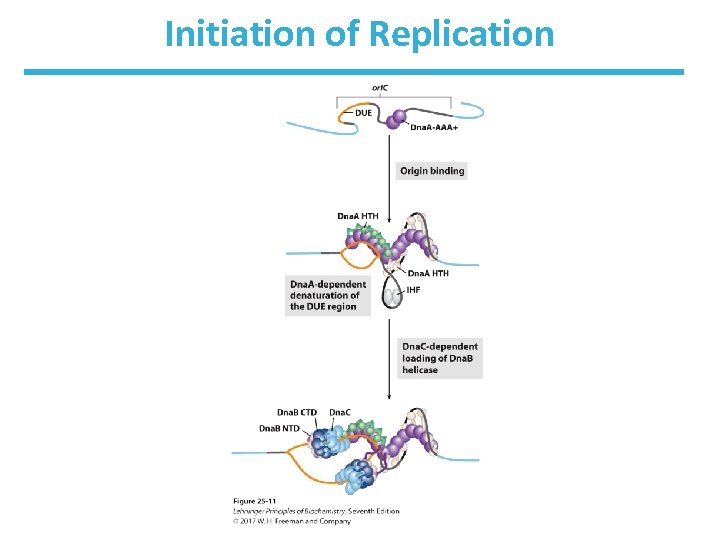
Initiation of Replication in E. Coli • Begins at the ori. C site (245 bp in length) • Contains highly conserved sequence elements – five repeats of a 9 -bp sequence (R sites) that form binding site for initiator protein Dna. A – A = T-rich region (DNA unwinding element (DUE)) – additional sites include: • Dna. A (I sites), • IHF (integration host factor) • FIS (factor for inversion stimulation)
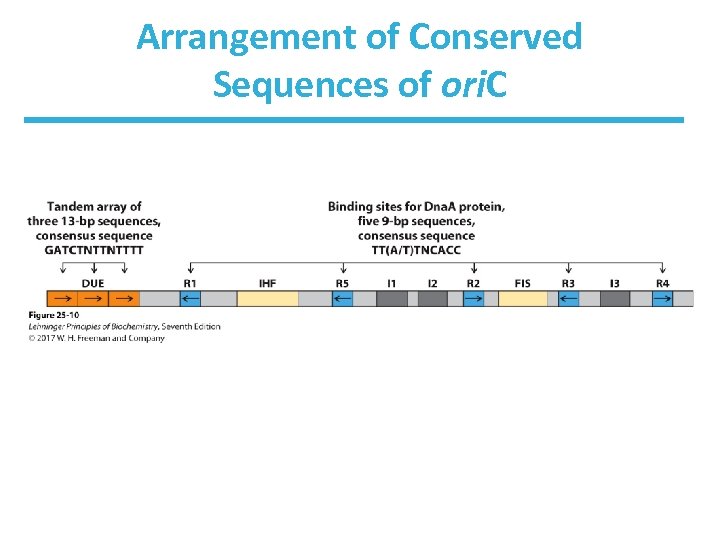
Arrangement of Conserved Sequences of ori. C
Requirements of Initiation of Replication in E. Coli • Requires at least 10 different proteins • See Table 25 -3 • Goal: open the helix, form prepriming complex
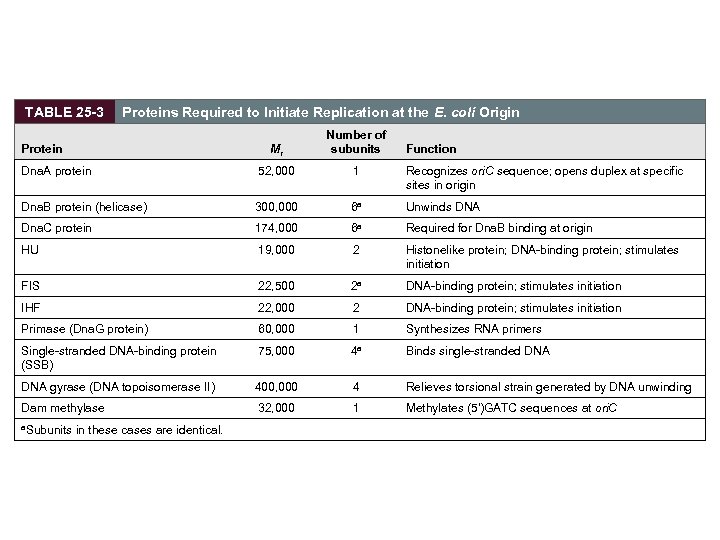
TABLE 25 -3 Proteins Required to Initiate Replication at the E. coli Origin Mr Number of subunits Dna. A protein 52, 000 1 Recognizes ori. C sequence; opens duplex at specific sites in origin Dna. B protein (helicase) 300, 000 6 a Unwinds DNA Dna. C protein 174, 000 6 a Required for Dna. B binding at origin HU 19, 000 2 Histonelike protein; DNA-binding protein; stimulates initiation FIS 22, 500 2 a DNA-binding protein; stimulates initiation IHF 22, 000 2 DNA-binding protein; stimulates initiation Primase (Dna. G protein) 60, 000 1 Synthesizes RNA primers Single-stranded DNA-binding protein (SSB) 75, 000 4 a Binds single-stranded DNA gyrase (DNA topoisomerase II) 400, 000 4 Relieves torsional strain generated by DNA unwinding Dam methylase 32, 000 1 Methylates (5')GATC sequences at ori. C Protein a. Subunits in these cases are identical. Function
Dna. A Proteins Bind at R and I Sites in ori. C • Dna. A proteins are ATPases. • Eight Dna. As bind to R and I sites. • DNA wraps around the complex, forming a positive supercoil. • Strain leads to denaturation of nearby DUE sites. • Associated proteins facilitate DNA bending.
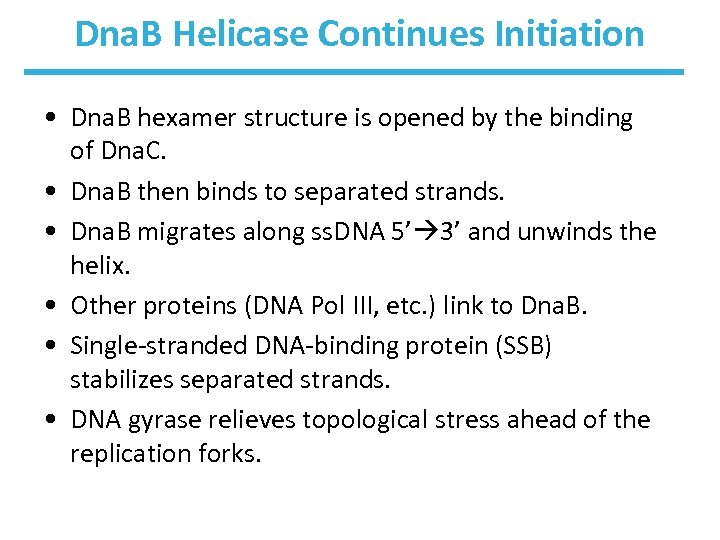
Dna. B Helicase Continues Initiation • Dna. B hexamer structure is opened by the binding of Dna. C. • Dna. B then binds to separated strands. • Dna. B migrates along ss. DNA 5’ 3’ and unwinds the helix. • Other proteins (DNA Pol III, etc. ) link to Dna. B. • Single-stranded DNA-binding protein (SSB) stabilizes separated strands. • DNA gyrase relieves topological stress ahead of the replication forks.
Initiation of Replication
Initiation Occurs Once Per Cell Cycle • Hda binds to subunits of DNA Pol III and stimulates hydrolysis of Dna. A’s ATP. – Hda is homologous to Dna. A • Dna. A complex then dissociates. • ADP dissociates. • ATP rebinds to Dna. A to stimulate all over again. • Time scale: 20– 40 mins
Regulation of Replication Initiation via Methylation • After replication, ori. C is hemimethylated by Dam methylase. – Dam = DNA adenine methylase – methylates N 6 of A in the palindromic GATC sequences • Hemimethylated ori. C sequences interact with the plasma membrane (uses protein Seq. A). • After a period, Seq. A dissociates, ori. C sequences are released from membrane. • Dam methylase fully methylates DNA to allow new Dna. A to bind.
Elongation of the Leading Strand • Straightforward approach • Primase (Dna. G) makes RNA primer (10– 60 nt). – The Dna. G primase interacts with Dna. B helicase, but primase moves in the opposite direction to helicase. • DNA Pol III adds nucleotides to the 3’ end of the strand. – Pol III is linked to Dna. B, which is tethered to the opposite DNA strand (3’ 5’). – ~1, 000– 2, 000 nt/sec
Elongation of the Lagging Strand • As in leading strand synthesis, primase makes RNA primer and DNA Pol III adds nucleotides. • Synthesis proceeds with new nucleotides adding on to the 3’ end of the new strand. • This strand is elongated away from the replication fork. • One asymmetric DNA Pol III dimer complex synthesizes both strands! – How? The DNA of the lagging strand loops around.
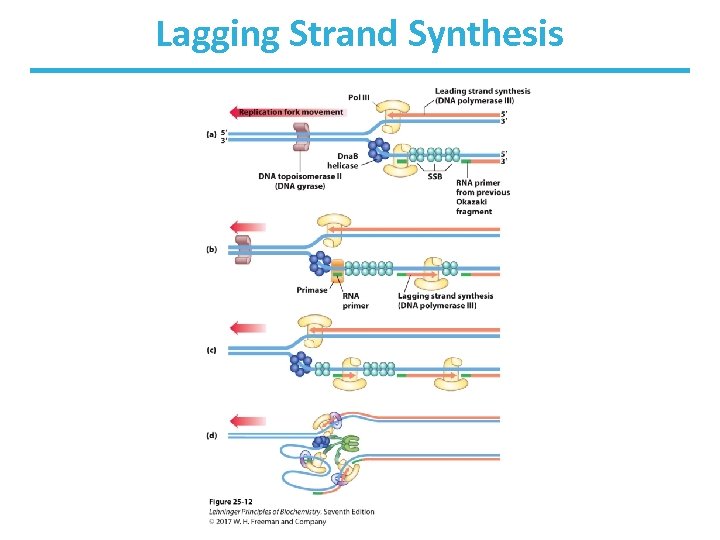
Lagging Strand Synthesis
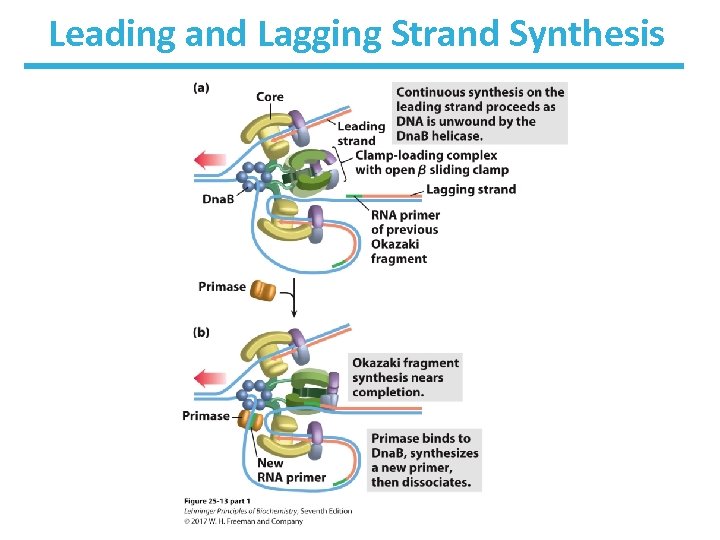
Leading and Lagging Strand Synthesis
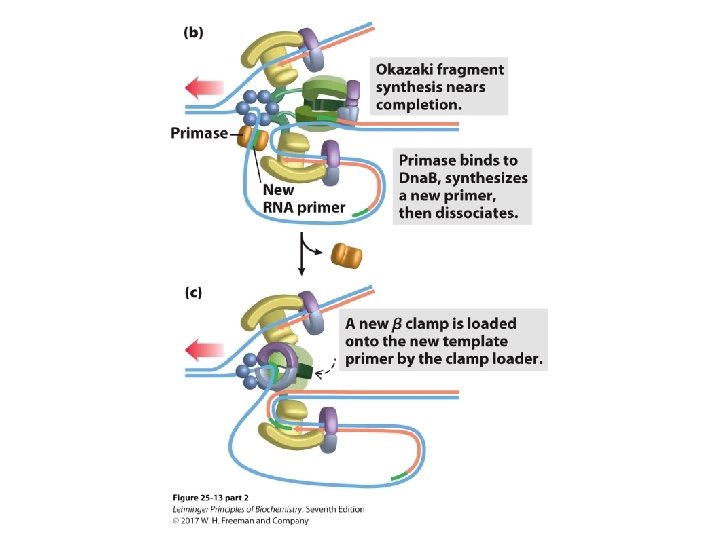
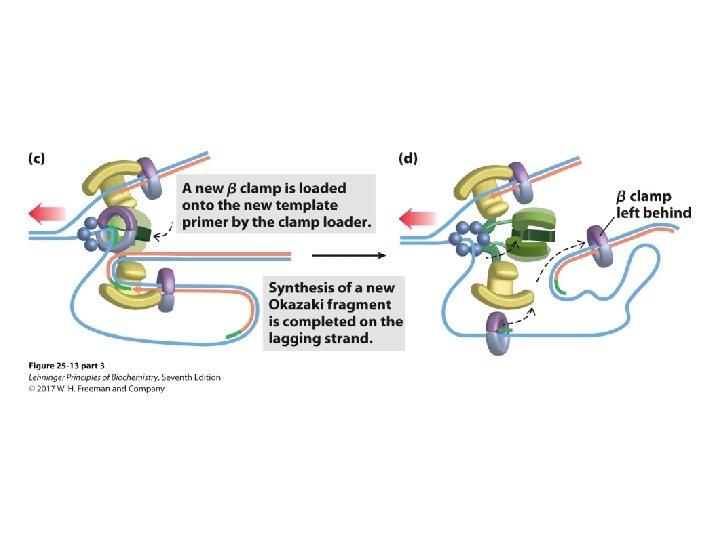
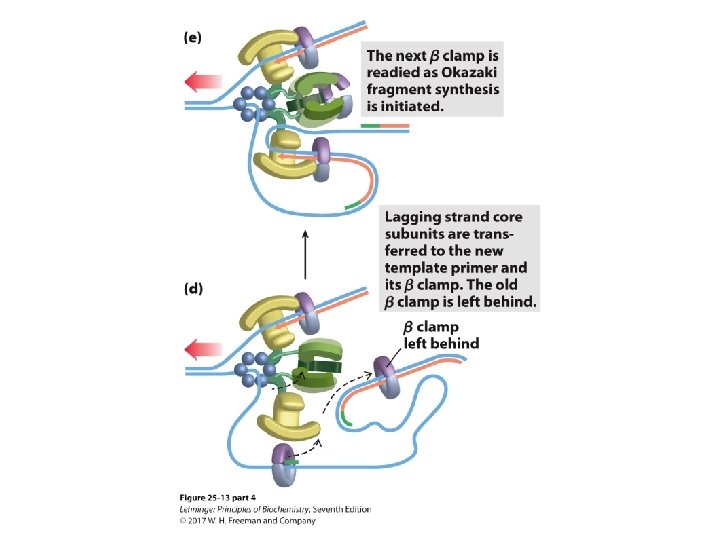
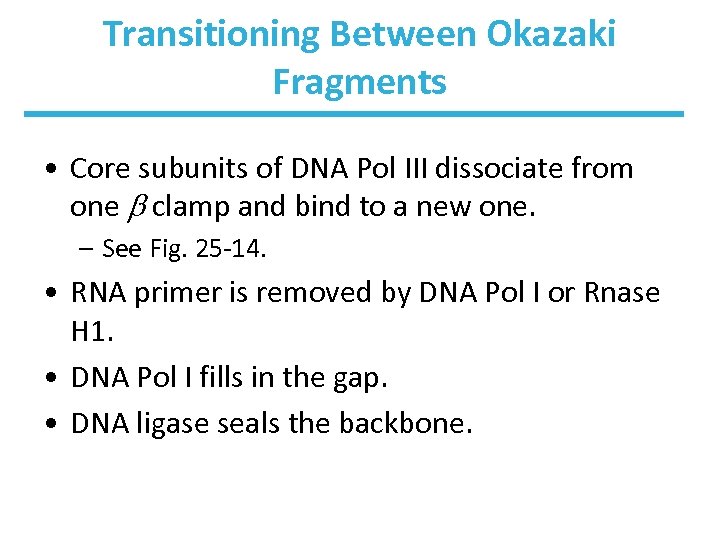
Transitioning Between Okazaki Fragments • Core subunits of DNA Pol III dissociate from one clamp and bind to a new one. – See Fig. 25 -14. • RNA primer is removed by DNA Pol I or Rnase H 1. • DNA Pol I fills in the gap. • DNA ligase seals the backbone.
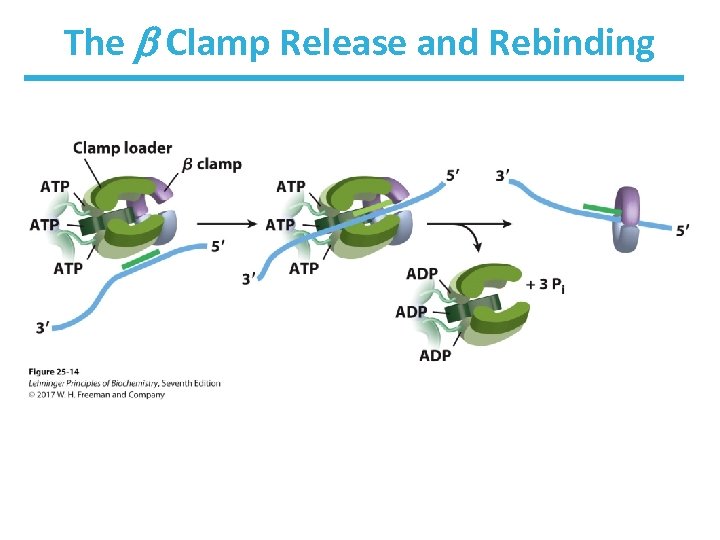
The Clamp Release and Rebinding
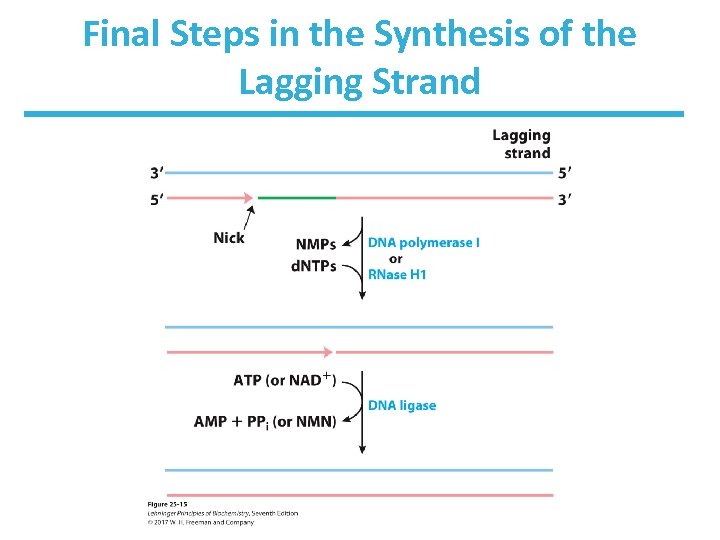
Final Steps in the Synthesis of the Lagging Strand
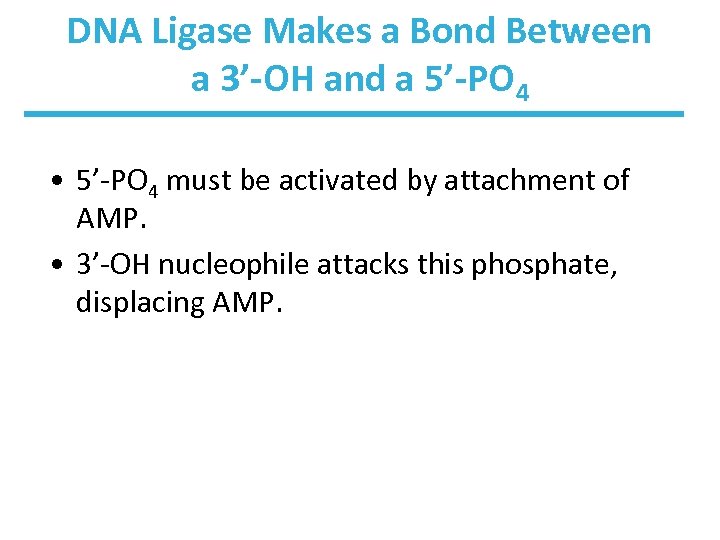
DNA Ligase Makes a Bond Between a 3’-OH and a 5’-PO 4 • 5’-PO 4 must be activated by attachment of AMP. • 3’-OH nucleophile attacks this phosphate, displacing AMP.
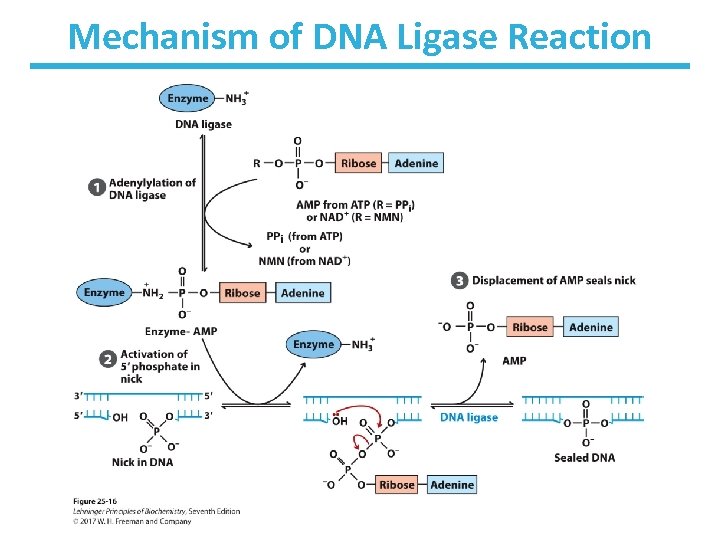
Mechanism of DNA Ligase Reaction
Third Stage of Replication: Termination • Replication forks meet at a region with 20 bp sequences Ter (Ter. A-Ter. F). – Ter sites found near each other but in opposite directions – Create a site that replication forks cannot leave • Ter is also a binding site for the protein Tus. – Tus = terminus utilization sequence – causes a replication fork to stop
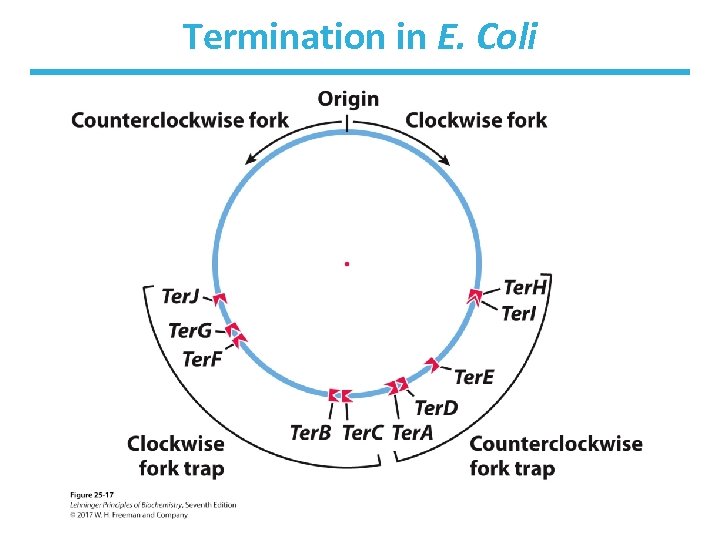
Termination in E. Coli
Replication in Eukaryotes • More complex than bacteria • Yeast have ~400 well-defined origins – called autonomously replicating sequences (ARS) or replicators • Entire genome replicated 1 X/cycle – regulation due to cyclin proteins and cyclindependent kinases (CDKs) – cyclins ubiquinated for proteolytic destruction at the end of the M (mitosis) phase
Initiation of Replication • Requires a prereplicative complex (pre-RCs) • The origin recognition complex (ORC) loads a helicase onto the DNA. – ORC functions like bacterial Dna. A. • Helicase is a hexamer of mini-chromosome maintenance proteins (MCM 2 -7). – MCM 2 -7 function like bacterial Dna. B helicase.
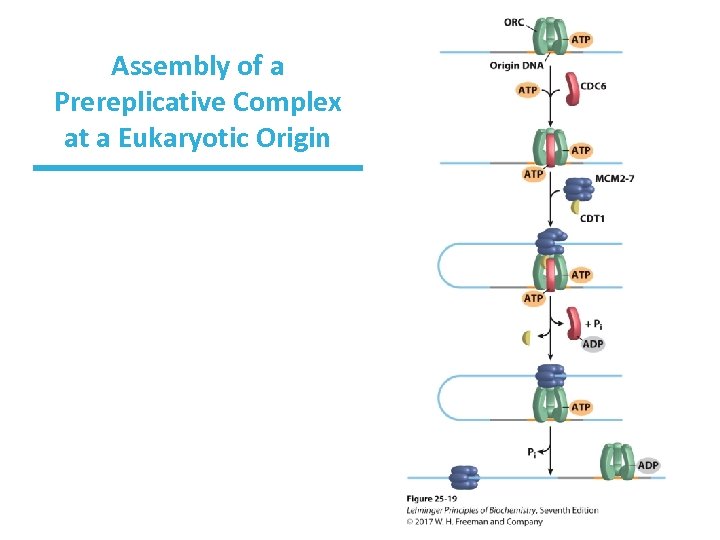
Assembly of a Prereplicative Complex at a Eukaryotic Origin
Eukaryotic Rate of Replication • Occurs more slowly than E. coli does • Synthesis ~50 nucleotides/sec – (~1/20 th the rate seen in E. coli) • Compensated by origins every 30– 300 kb
Multiple DNA Polymerases in Eukaryotic Nuclear Replication • DNA Pol - polymerase/primase activity – has primase activity in one subunit and polymerization in another – does not have 3’ 5’ proofreading • DNA Pol (lagging strand) and DNA Pol (leading strand) - associated with PCNA (proliferating cell nuclear antigen-protein) – both highly processive; comparable to bacterial DNA Pol III – both 3’ 5’ proofreading
Termination of Replication • Synthesis of specialized structures known as telomeres are found at the linear ends of the nuclear chromosomes. • The enzyme telomerase uses RNA and is therefore discussed in Chapter 26.
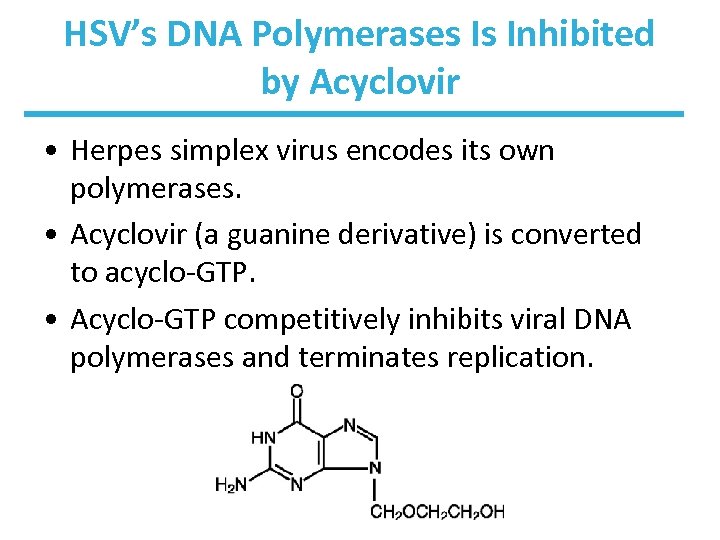
HSV’s DNA Polymerases Is Inhibited by Acyclovir • Herpes simplex virus encodes its own polymerases. • Acyclovir (a guanine derivative) is converted to acyclo-GTP. • Acyclo-GTP competitively inhibits viral DNA polymerases and terminates replication.
DNA Repair and Mutations • Chemical reactions and some physical processes constantly damage genomic DNA. – The majority are corrected using the undamaged strand as a template. – Some base changes escape repair, and an incorrect base serves as a template in replication. – The daughter DNA carries a changed sequence in both strands. • Accumulation of mutations in eukaryotic cells is strongly correlated with cancer; most carcinogens are also mutagens. • There are thousands of lesions/day (unrepaired DNA damage) but only 1/1, 000 become a mutation, thanks to DNA repair. • The human genome contains genes for > 130 repair proteins.
Vocabulary of DNA Lesions • Lesion = DNA damage • If unrepaired, lesion becomes mutation – Mutations can be substitutions (point mutations), deletions, additions • Silent mutation―has ~no effect on gene function or affects a nonessential region of the DNA
The Ames Test • Developed by Bruce Ames • Indicates the mutagenic potential of a compound • Uses Salmonella strain with a mutation that makes bacterium unable to synthesize His • Add compound to plate of Salmonella, see if it grows in His-free medium – Colonies (+ test) indicates the compound mutated the Salmonella, restored ability to synthesize His
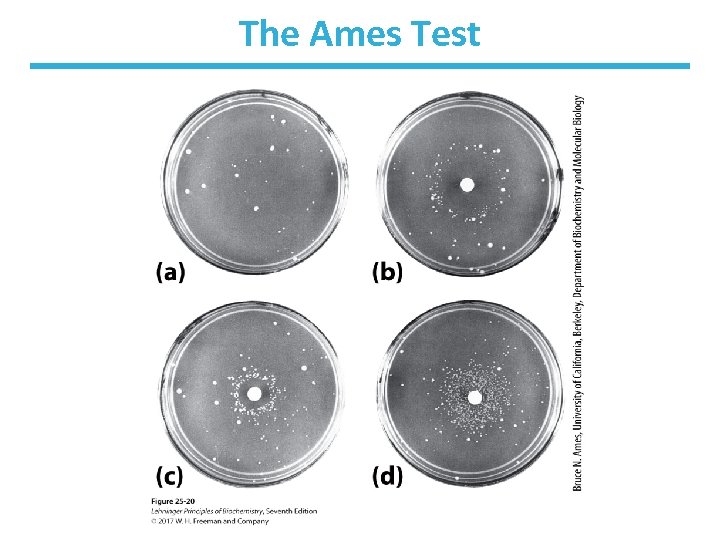
The Ames Test
Types of DNA Damage • Mismatches arise from occasional incorporation of incorrect nucleotides. • Abnormal bases arise from spontaneous deamination, chemical alkylation, or exposure to free radicals. • Pyrimidine dimers form when DNA is exposed to UV light. • Backbone lesions occur from exposure to ionizing radiation and free radicals.
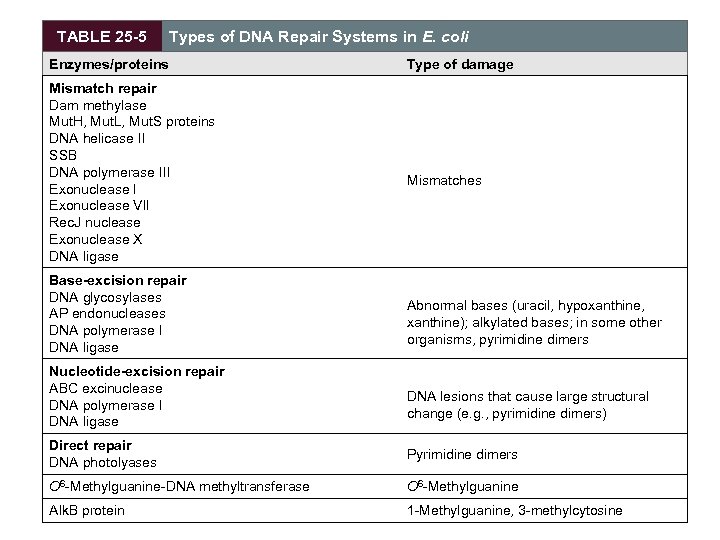
TABLE 25 -5 Types of DNA Repair Systems in E. coli Enzymes/proteins Mismatch repair Dam methylase Mut. H, Mut. L, Mut. S proteins DNA helicase II SSB DNA polymerase III Exonuclease VII Rec. J nuclease Exonuclease X DNA ligase Base-excision repair DNA glycosylases AP endonucleases DNA polymerase I DNA ligase Nucleotide-excision repair ABC excinuclease DNA polymerase I DNA ligase Type of damage Mismatches Abnormal bases (uracil, hypoxanthine, xanthine); alkylated bases; in some other organisms, pyrimidine dimers DNA lesions that cause large structural change (e. g. , pyrimidine dimers) Direct repair DNA photolyases Pyrimidine dimers O 6 -Methylguanine-DNA methyltransferase O 6 -Methylguanine Alk. B protein 1 -Methylguanine, 3 -methylcytosine
Mismatch Repair Relies on Methylation How do repair enzymes “know” which strand is the correct one? • In E. coli, the parent strand is methylated. • Dam methylase inserts CH 3 at adenines in the GATC sequence. • Following a short period of time, the daughter strand is then methylated.
Methylation of DNA and Repair • The newly synthesized strand is unmethylated for a short period after synthesis. • Any replication errors must reside in the unmethylated strand. • The methyl-directed mismatch repair system will cleave the unmethylated strand in the initial part of the repair process.
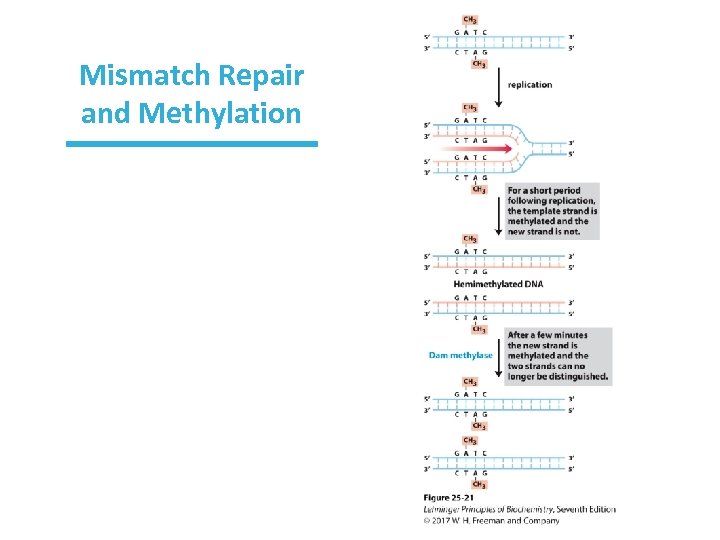
Mismatch Repair and Methylation
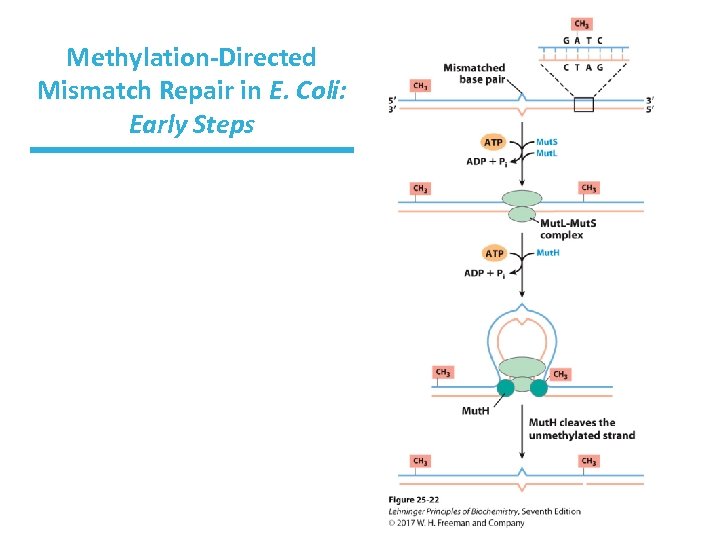
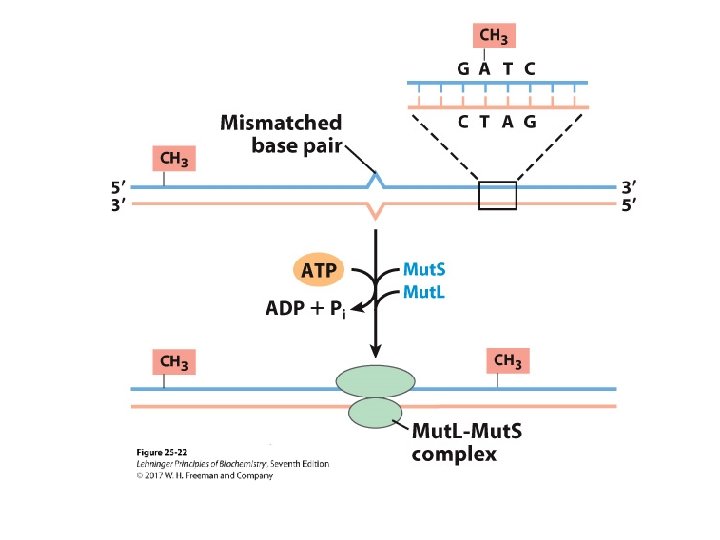
Mechanism of Mismatch Recognition and Repair in E. coli • Mut. L and Mut. S proteins recognize methylated GATC. • Mut. H binds to Mut. L-Mut. S-DNA complex, making a DNA loop. • DNA strands thread through the complex until methylated GATC is encountered. – mismatch could be 1, 000 bp away from GATC
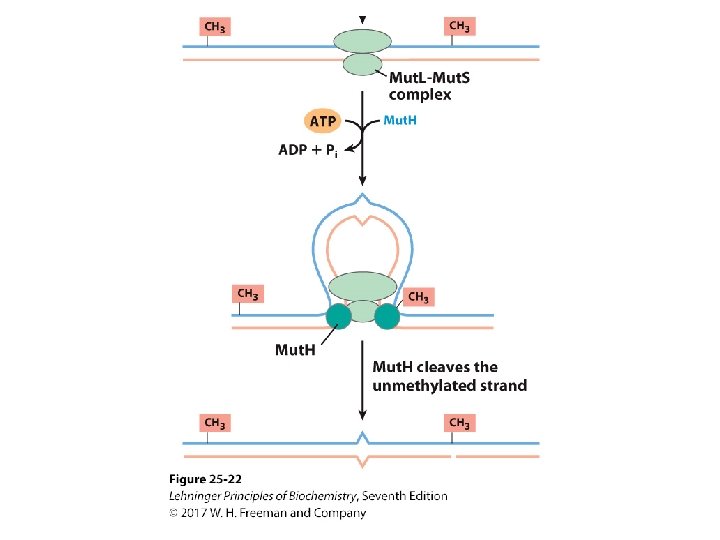
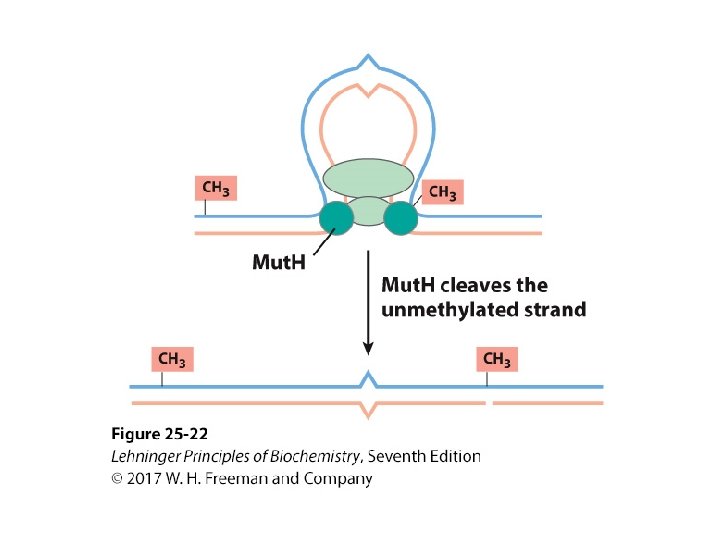
Mechanism of Mismatch Repair in E. Coli • Mut. H cleaves the nonmethylated DNA strand on the 5’-side of the G. • DNA unwinds and is degraded 3’ 5’. – Helicase II (Uvr. D helicase), SSB, and exonucleases work to degrade the nonmethylated DNA toward the mismatch. • The removed sequence is replaced using DNA Pol III and DNA ligase.
Methylation-Directed Mismatch Repair in E. Coli: Early Steps
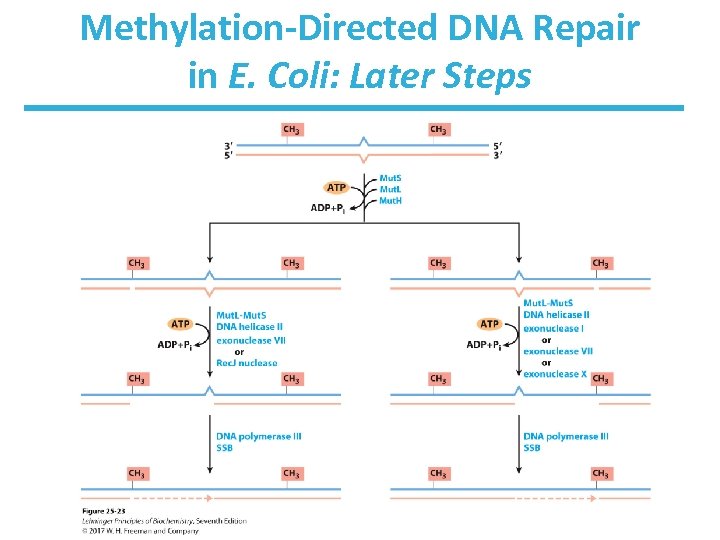
Methylation-Directed DNA Repair in E. Coli: Later Steps
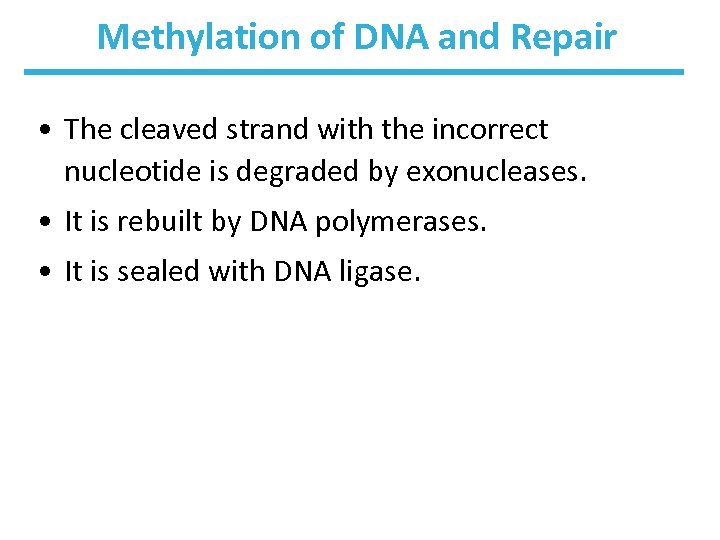
Methylation of DNA and Repair • The cleaved strand with the incorrect nucleotide is degraded by exonucleases. • It is rebuilt by DNA polymerases. • It is sealed with DNA ligase.
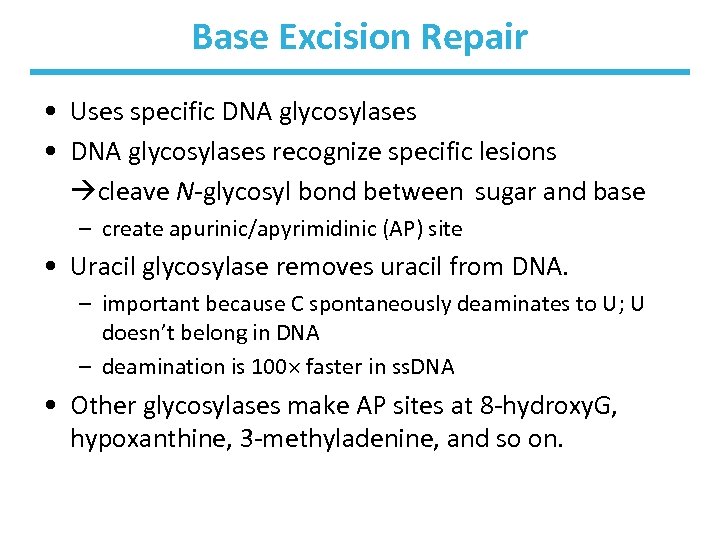
Base Excision Repair • Uses specific DNA glycosylases • DNA glycosylases recognize specific lesions cleave N-glycosyl bond between sugar and base – create apurinic/apyrimidinic (AP) site • Uracil glycosylase removes uracil from DNA. – important because C spontaneously deaminates to U; U doesn’t belong in DNA – deamination is 100 faster in ss. DNA • Other glycosylases make AP sites at 8 -hydroxy. G, hypoxanthine, 3 -methyladenine, and so on.
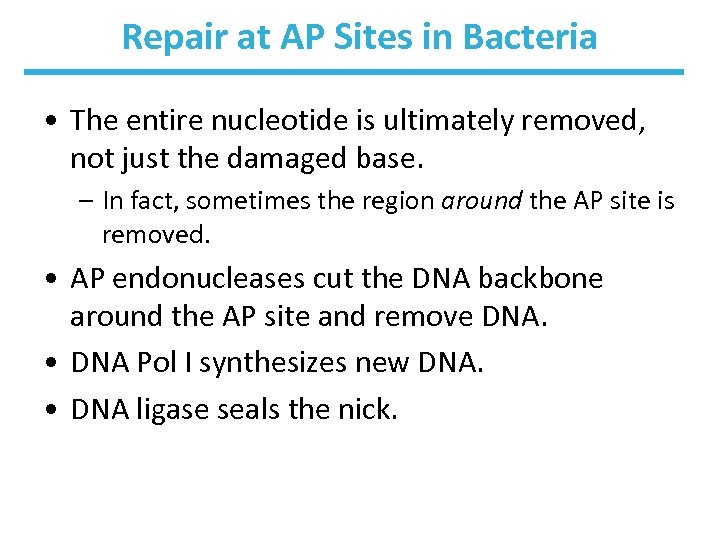
Repair at AP Sites in Bacteria • The entire nucleotide is ultimately removed, not just the damaged base. – In fact, sometimes the region around the AP site is removed. • AP endonucleases cut the DNA backbone around the AP site and remove DNA. • DNA Pol I synthesizes new DNA. • DNA ligase seals the nick.
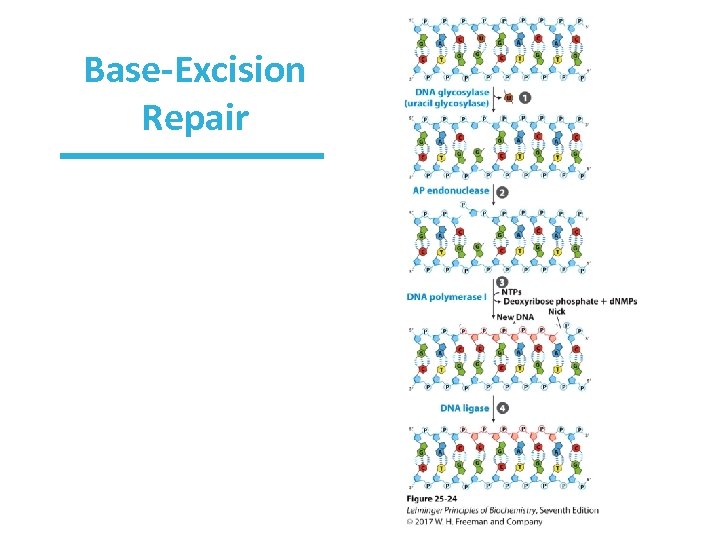
Base-Excision Repair
Large Distortions in DNA Are Repaired by Nucleotide Excision • Lesions include: – pyrimidine dimers (formed from UV light) – 6, 4 -photoproducts (from UV light) – benzo[a]pyreneguanine (from cigarette smoke) • Pathway involves removal of a DNA segment by excinucleases.
Nucleotide Excision Repair in Bacteria Uses the ABC Excinuclease • Excinucleases cleave DNA backbone in two places. • ABC excinuclease contains Uvr. A, Uvr. B, and Uvr. C. – hydrolyze fifth bond on 3’-side of the lesion and eighth bond on 5’-side – remove 12– 13 nucleotides • DNA Pol I and DNA ligase to replace the DNA and seal the gap
Nucleotide Excision Repair (NER) in Eukaryotes • Hydrolyze sixth bond on 3’-side and twentysecond bond on 5’-side of lesion – remove 27– 29 nucleotides • In humans, gap filled using DNA polymerase and DNA ligase
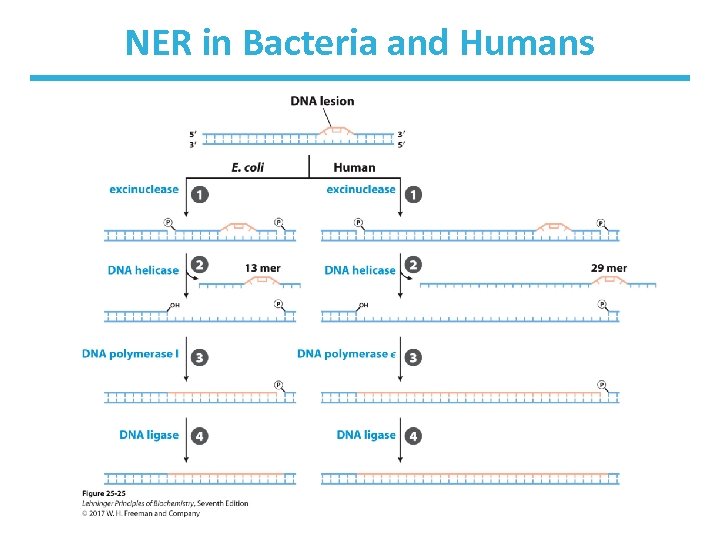
NER in Bacteria and Humans

Direct Repair • Photolyases use light energy to repair pyrimidine dimers. – enzymes not found in humans and placental mammals • O 6 -methylguanine-DNA methyltransferase repairs methylated guanine. • Alk. B demethylates 1 -methyladenine and -methylcytosine. 3
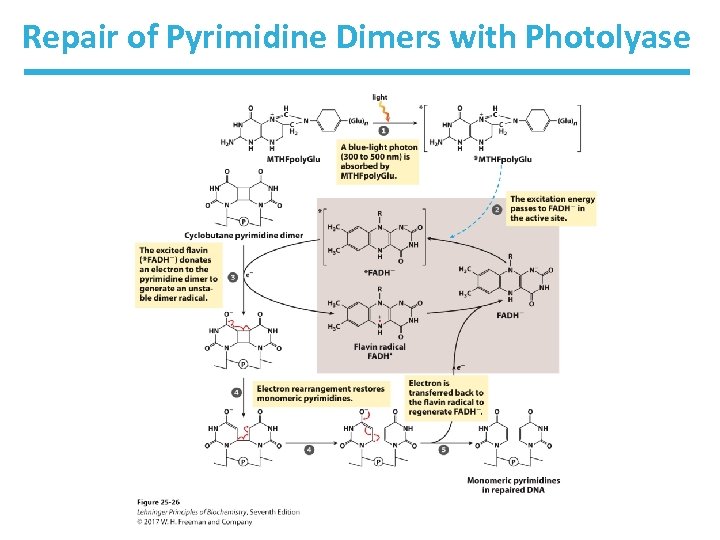
Repair of Pyrimidine Dimers with Photolyase
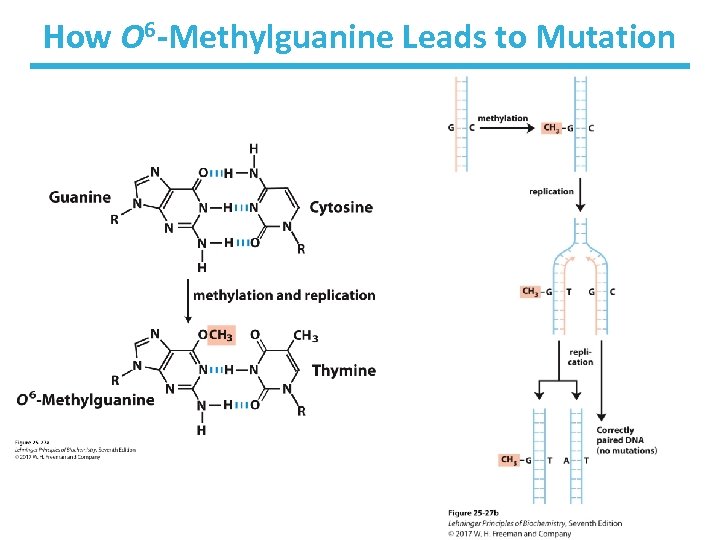
How O 6 -Methylguanine Leads to Mutation
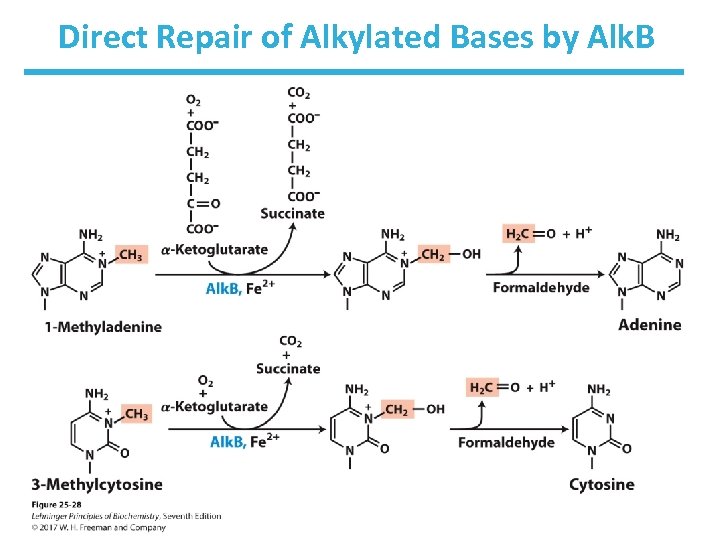
Direct Repair of Alkylated Bases by Alk. B
What Happens When There Is No Undamaged DNA to Use as a Template for Repair? • No template with dsb, cross-links, or damage in ss. DNA – Unrepaired lesion cause replication fork to stall • Repair using 1. another chromosome as template (recombination) 2. error-prone translesion synthesis (TLS)
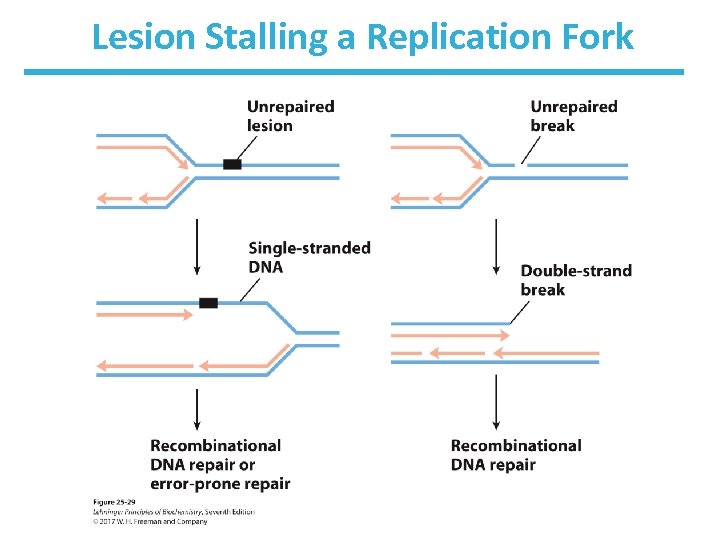
Lesion Stalling a Replication Fork
Error-Prone TLS in Bacteria • TLS is part of the “SOS response. ” – response when DNA damage is extensive • SOS proteins include Uvr. A, Uvr. B + Umu. C, and Umu. D. – Umu = Unmutable (because lack of Umu genes abolishes the pathway) • Cleaved Umu. D’ and Umu. C bind with Rec. A to create DNA Pol V. – This polymerase can process past the damaged area.

Why Have Error-Prone Repair? • It kills some cells, mutates DNA in others. • But…some cells survive.
TLS Polymerases Abound in Mammals • Most recognize specific types of damage and have appropriate response. • Example: DNA Pol (eta) assists when a T-T dimer halts a replication fork. • Most TLS polymerases are limited to short regions of DNA, minimizing mutagenic potential.
DNA Recombination • Segments of DNA can rearrange their location – within a chromosome – from one chromosome to another • Such recombination is involved in many biological processes. – repair of DNA – segregation of chromosomes during meiosis – enhancement of genetic diversity • In sexually reproducing organisms, recombination and mutations are two driving forces of evolution. • Recombination of co-infecting viral genomes may enhance virulence and provide resistance to antivirals.
Three Classes of Recombination • Homologous/general recombination – exchange between two DNAs that share an extended region of similar sequence • Site-specific recombination – exchange only at a particular sequence • DNA transposition – “jumping genes” – short DNAs that can move from one chromosome to another
Three Functions of Homologous Recombination 1. Assists in DNA repair 2. Links sister chromosomes to properly segregate them between self and daughter cells 3. Source of DNA exchange and therefore genetic diversity
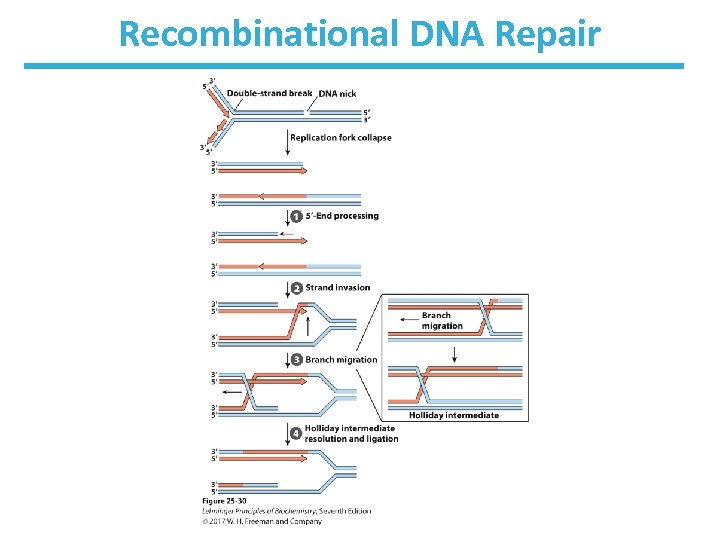
Homologous Recombination in Bacteria • See Fig. 25 -30 • Replication fork encounters damage in template strand • Replication fork collapses due to creation of a double-strand break (DSB)
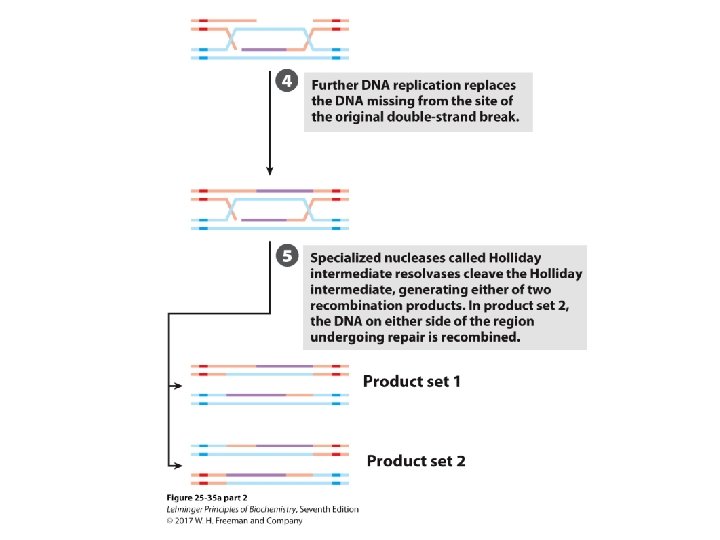
Bacterial Recombination (cont. ) – leaves two parts of a strand to be processed • 5’-ending strand is degraded. • 3’-single-stranded extension is bound by a recombinase, pairs with complementary sequence in intact duplex DNA, and “invades” the duplex. • creates branched structure of three strands. • Branch moves to create X-like structure known as “Holliday junction” or “Holliday intermediate. ” • A nuclease and ligase restore the structure of the replication fork.
Recombinational DNA Repair
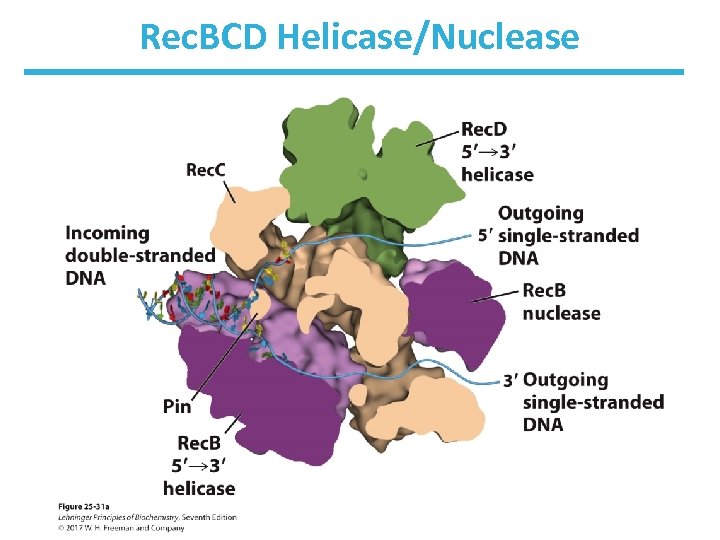
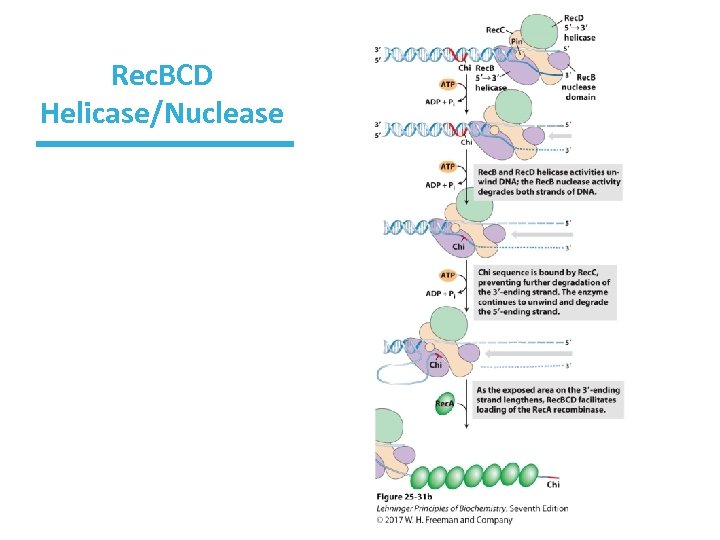
Rec. BCD Enzyme • A nuclease/helicase processes the DNA ends. • It binds to a broken end and then unwinds and hydrolyzes the DNA. • Rec. B moves along one strand 3’ 5’. • Rec. D moves along the other strand 5’ 3’. • The Rec. C subunit binds to the chi sequence (GCTGGTGG) and alters the activity of the enzyme complex.
Rec. BCD Helicase/Nuclease
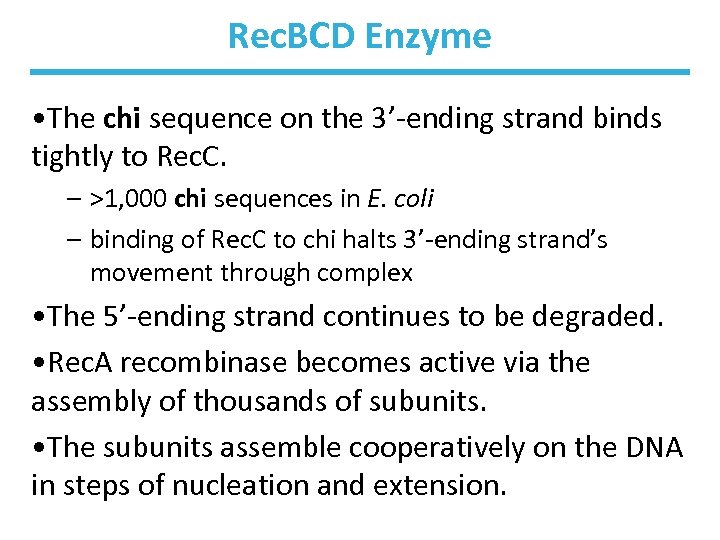
Rec. BCD Enzyme • The chi sequence on the 3’-ending strand binds tightly to Rec. C. – >1, 000 chi sequences in E. coli – binding of Rec. C to chi halts 3’-ending strand’s movement through complex • The 5’-ending strand continues to be degraded. • Rec. A recombinase becomes active via the assembly of thousands of subunits. • The subunits assemble cooperatively on the DNA in steps of nucleation and extension.
Rec. BCD Helicase/Nuclease
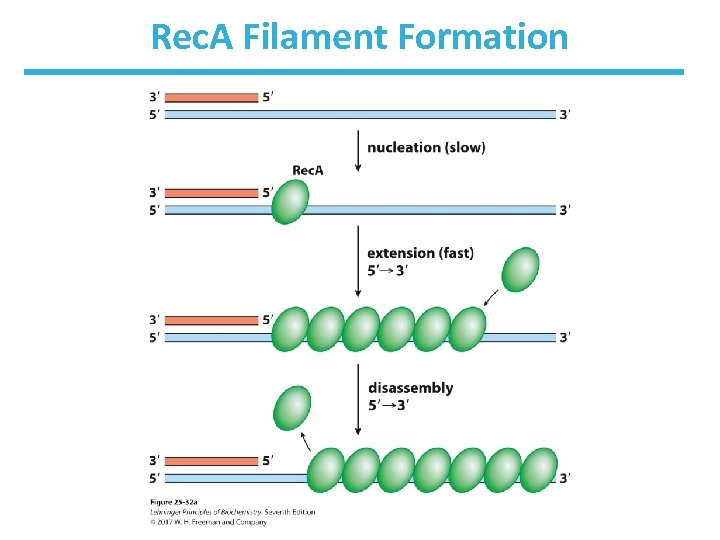
Rec. A Filament Formation
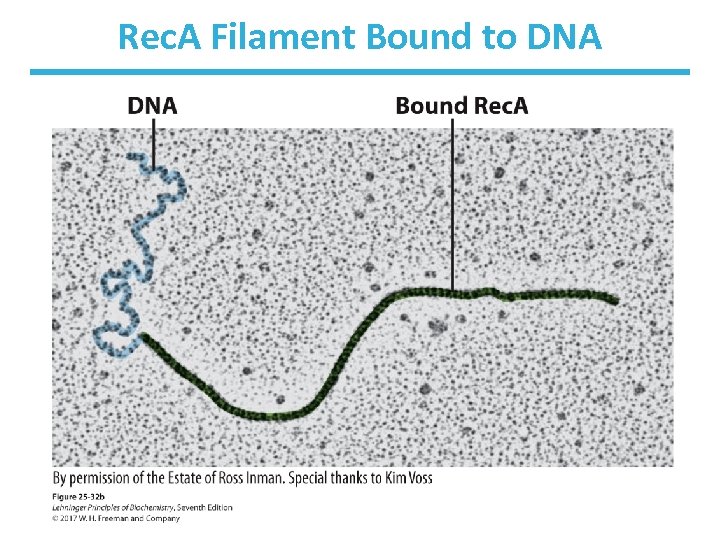
Rec. A Filament Bound to DNA
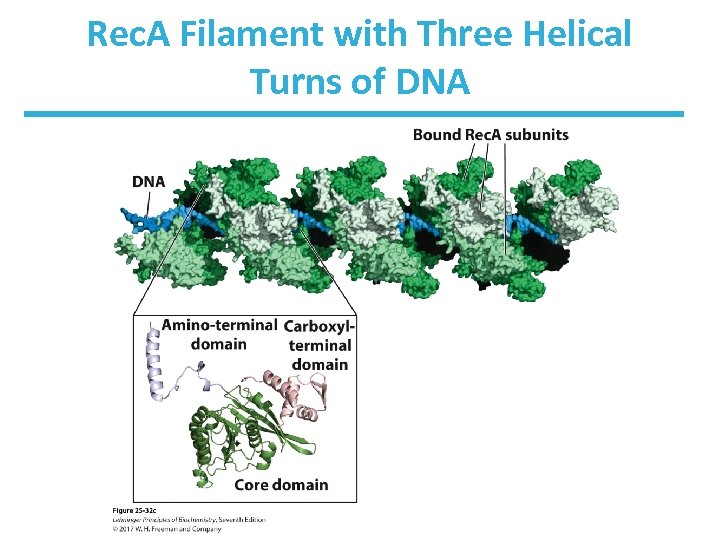
Rec. A Filament with Three Helical Turns of DNA
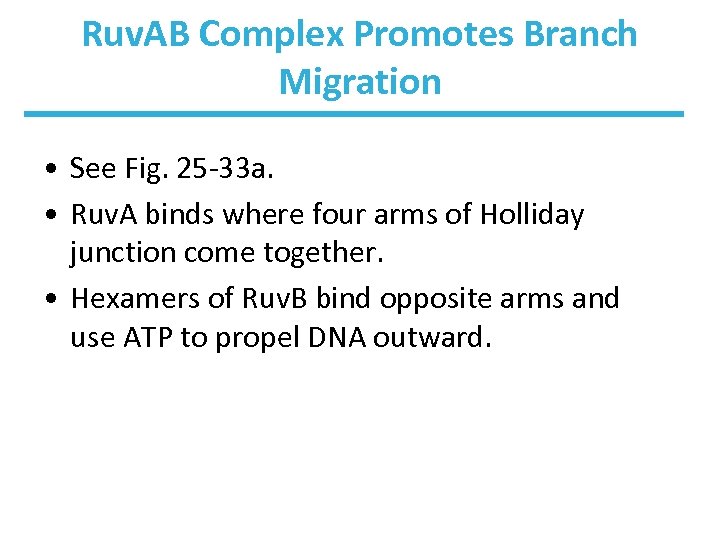
Ruv. AB Complex Promotes Branch Migration • See Fig. 25 -33 a. • Ruv. A binds where four arms of Holliday junction come together. • Hexamers of Ruv. B bind opposite arms and use ATP to propel DNA outward.
Holliday Junction Is Cleaved by Ruv. C • Ruv. C is a nuclease. • Binds to Ruv. AB complex. • It cleaves the Holliday intermediate on opposite sides. • DNA ligase seals the nick. • See Fig. 25 -33 b.
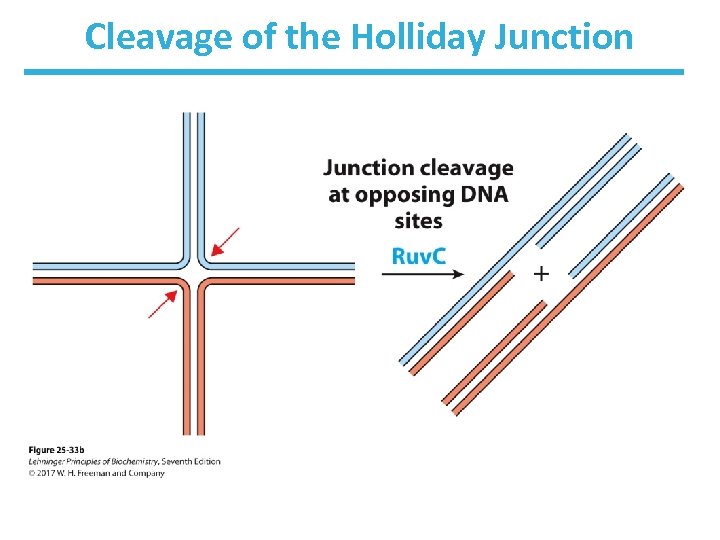
Cleavage of the Holliday Junction
Homologous Replication in Eukaryotes • Occurs frequently during meiosis • Meiosis: when diploid germ cells divide to produce haploid cells – It iinitially produces four homologous chromosomes (a tetrad). – The two pairs of sister chromatids are connected at centromeres. • Cells then divide twice – Homologous chromosomes are segregated into separate daughter cells at the end of meiosis I. – Sister chromatids are separated during meiosis II.
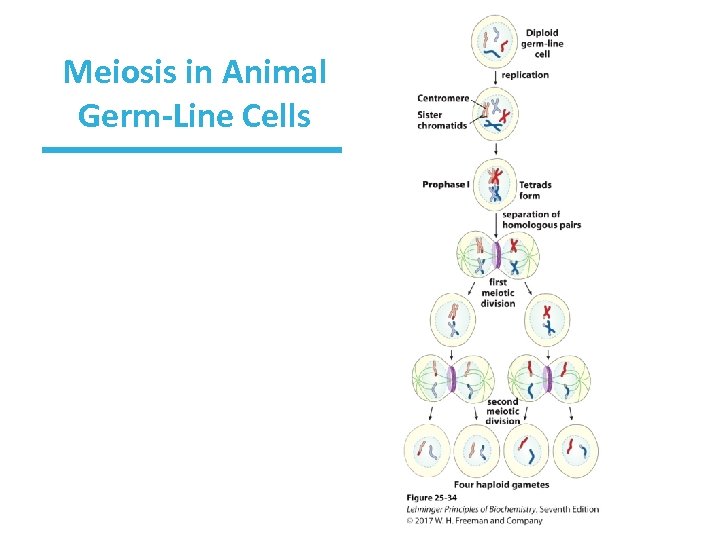
Meiosis in Animal Germ-Line Cells

Recombination Links Sister Chromatids Together • Recombination (crossing over) occurs in the first meiotic division. • Sequences called chiasmata are where the strand breaks and rejoining occurs. • This occurs more often at “hot spots. ” • This process not only aligns sister chromosomes for proper segregation, but it allows for exchange of genetic information, thus increasing genetic diversity. • In the second meiotic division, physical links created by cohesins align chromatids and help them segregate properly.
Model of Homologous Recombination in Meiosis • Chromosomes align (call them A and B) • Double-strand break occurs in chromosome A. – Exonuclease degrades dsb ends to make bigger gap. • 3’-ending strands of the gap “invade” chromosome B they pair with a complementary region on chromosome B. – This displaces a strand in chromosome B; it binds to chromosome A. – The displaced region of chromosome B (now paired with chromosome A) serves as the template to replace missing DNA.
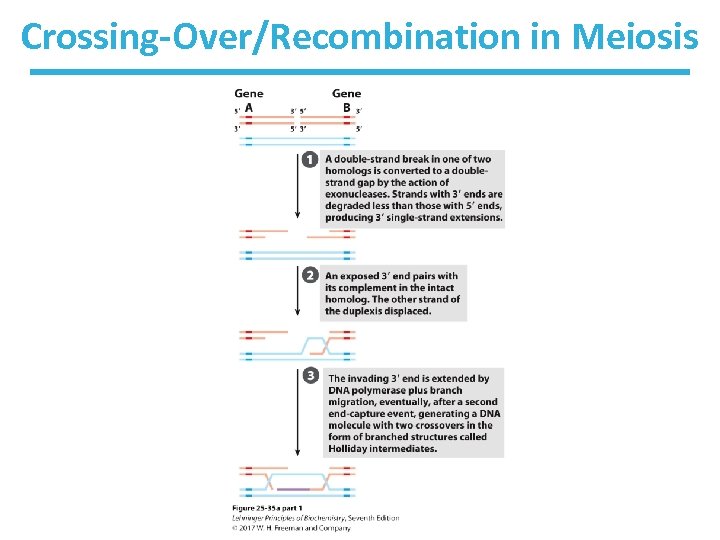
Crossing-Over/Recombination in Meiosis
Homolgous Recombination and Gene Mapping • In general, the frequency of homologous recombination between two points on a chromosome is roughly proportional to distance. • Allows position of genes (gene mapping) and linkage to be determined.
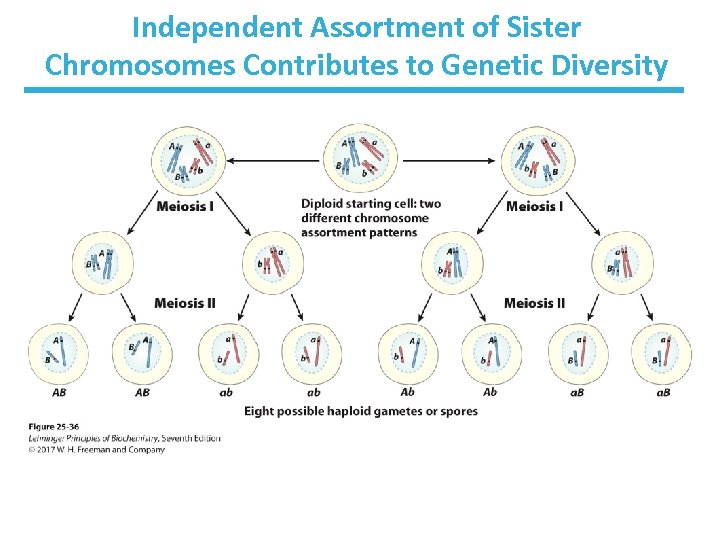
Independent Assortment of Sister Chromosomes Contributes to Genetic Diversity
Improper Segregation of Chromosomes • Aneuploidy = wrong # chromosomes • Improper alignment and/or recombination can result in gametes with no copies of a particular chromosomes or two copies. – Aneuploidy can lead to miscarriage or developmental disability. • It often occurs when a gamete with one copy (normal) joins with another gamete: – containing two copies trisomy • Trisomy of chromosome 21 is Down syndrome. – containing no copies monosomy • Monosomy of the X chromosome is Turner’s syndrome.

Differences in Male versus Female Meiosis Explain Aneuploidy • Males: Germ-line cells begin to undergo meiosis at puberty…process is rapid and frequent. • Females: There is a much longer-term process with an extended “suspended animation” phase. – Germ-line cells begin to undergo meiosis in the fetal stage…chromosomes align and undergo crossing-over, then the process stops for 13– 50 years until the egg cells mature (prophase I). – Disruptions to crossovers during this stage produce aneuploidy. – The likelihood of damage increases with age.
Nonhomologous End Joining (NHEJ) • Another way to repair DSB – not ideal but better than cell death • Broken chromosome ends are simply processed and ligated back together. • No conservation of DNA sequence • Ku 70 and Ku 80 proteins bind the DNA ends followed by other proteins—DNA-PKcs (kinase) and Artemis (nuclease). • DNA-PKcs autophosphorylates in several locations and phosphorylates Artemis.

Nonhomologous End Joining (NHEJ) • Artemis now removes 5’ or 3’ single-stranded nucleotides or hairpins that have formed. • Helicase separates the ends. • The remaining nick is sealed by a complex consisting of XRCC 4, XLF, and DNA ligase IV. • This form of repair typically occurs when the chromosomes are condensed and constrained, thus the ends are near each other.
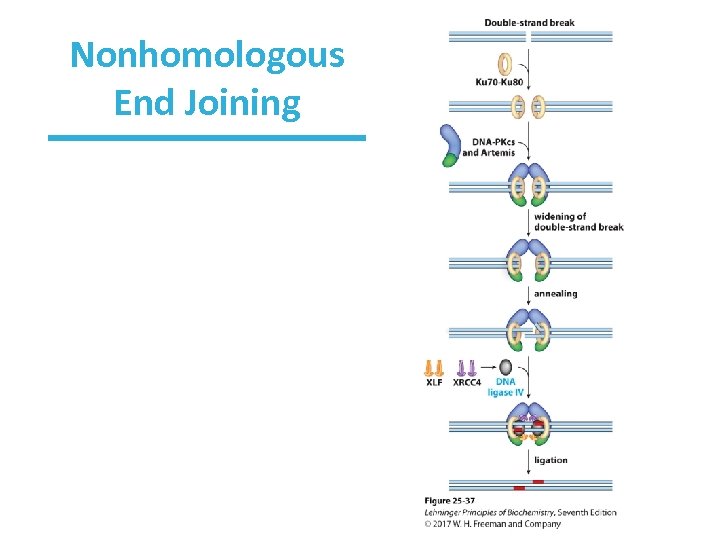
Nonhomologous End Joining
Site-Specific Recombination • It is limited to specific sequences. • It uses recombinase enzymes with either Tyr or Ser. • The recombinase makes covalent bond with DNA (phosphotyrosine bond, etc. ). • Cleaved strand joins new partners. • Exchange is reciprocal.
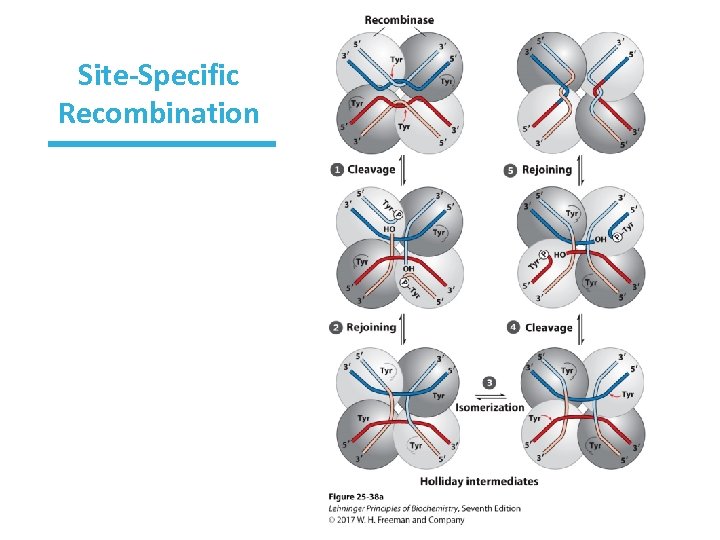
Site-Specific Recombination
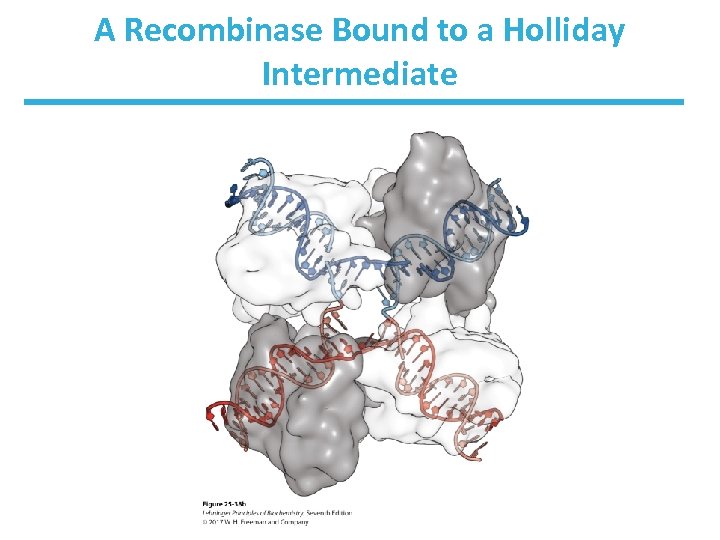
A Recombinase Bound to a Holliday Intermediate
Potential Outcomes of Site-Specific Recombination • Inversion or deletion – occurs if recombination sites are on the same DNA • Intermolecular recombination – occurs if recombination sites are on different DNAs • Insertion – occurs if DNA is circular
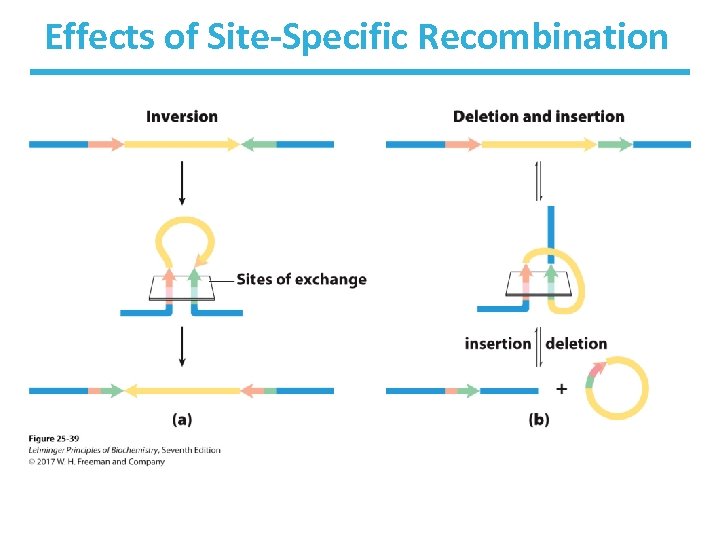
Effects of Site-Specific Recombination
Recombinational Repair of Circular DNA May Yield a Dimeric Genome • Resolution of Holliday junction can create dimeric DNAs. • A special site-specific recombination system converts dimer into monomers.
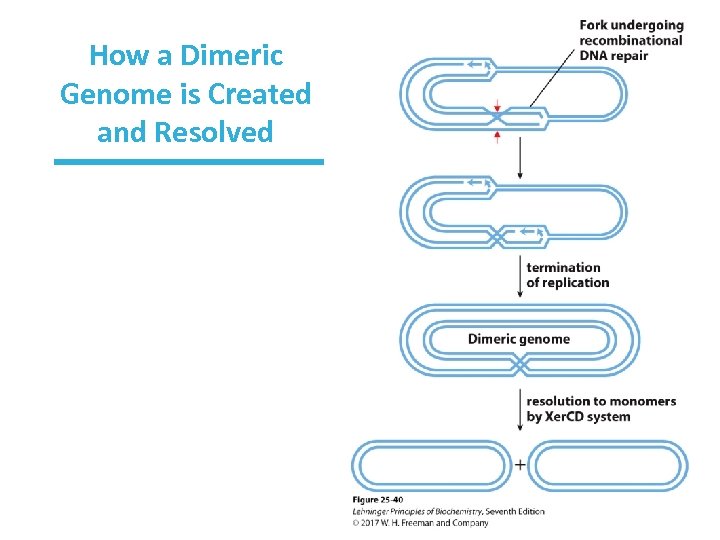
How a Dimeric Genome is Created and Resolved
Transposable Genetic Elements (Transposons) • Certain DNA segments can change their relative position (move to a region or a new chromosome). • They carry genes for transposases, but some can contain extra genes (e. g. , antibiotic resistance). • In bacteria, short (5– 10 bp) sequences act as binding sites for transposases. • These sites become duplicated. • See Fig. 25 -40.
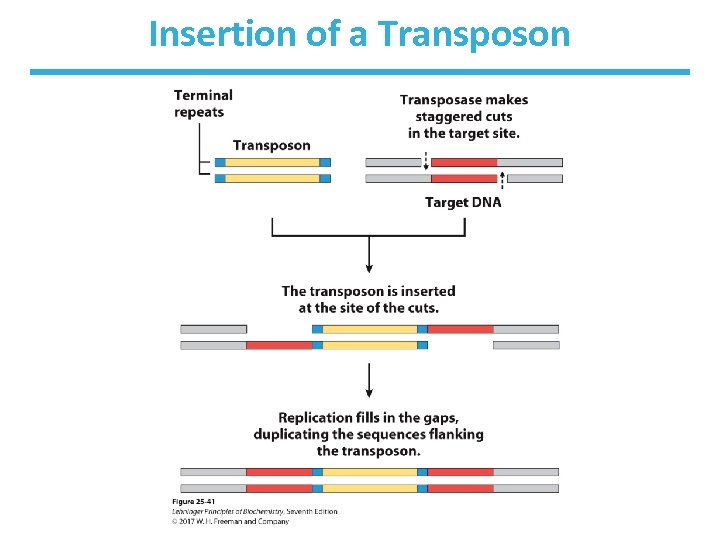
Insertion of a Transposon
Replicative Transposition • Previous (direct/simple) transposition: “cut and paste” • Replicative transposition: “copy and paste” • Entire transposon is replicated
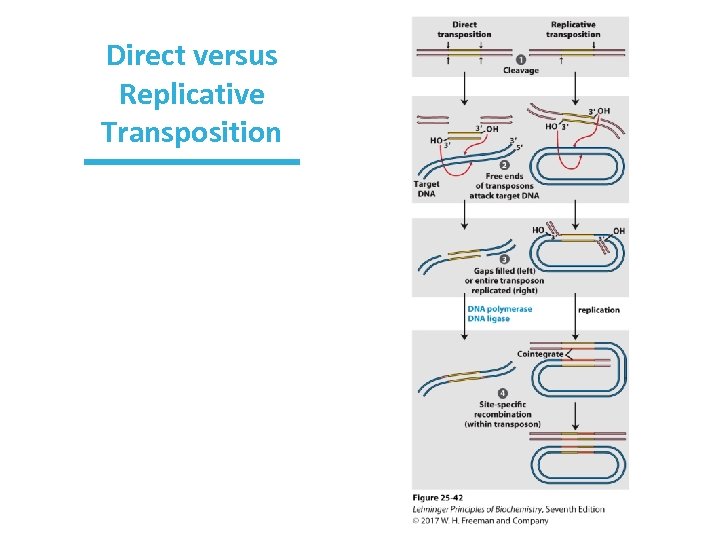
Direct versus Replicative Transposition
Immunoglobulin Genes Assemble by Recombination • The human genome contains ~20, 000 genes, yet millions of antibodies (immunoglobulins) can be created from the separate gene segment sets.
Immunoglobulin Genes Undergo Recombination • Structure: (example from Ig. G class) • Two heavy, two light polypeptide chains – Light chains can be kappa or gamma. • Each chain has variable (V) region and constant region. • Recombination between clusters of these genes produces different antibodies.
Kappa Chain V, J, and C Segments Undergo Recombination • Kappa chain gene has: – ~300 V (variable segments) – ~4 J (joining) segments – 1 C (constant) segment 300 4 = 1, 200 possible V-J arrangements • V-J junction sequence is not precise, producing more variation (2. 5 ): to 3, 000 possible combinations • Heavy chain and lambda chains have more combinations. • Ultimately > 1. 5 107 possible Ig. G molecules!
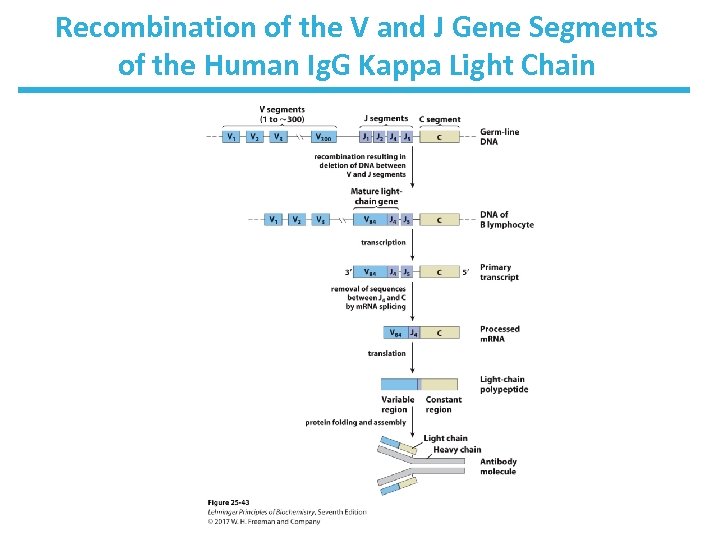
Recombination of the V and J Gene Segments of the Human Ig. G Kappa Light Chain
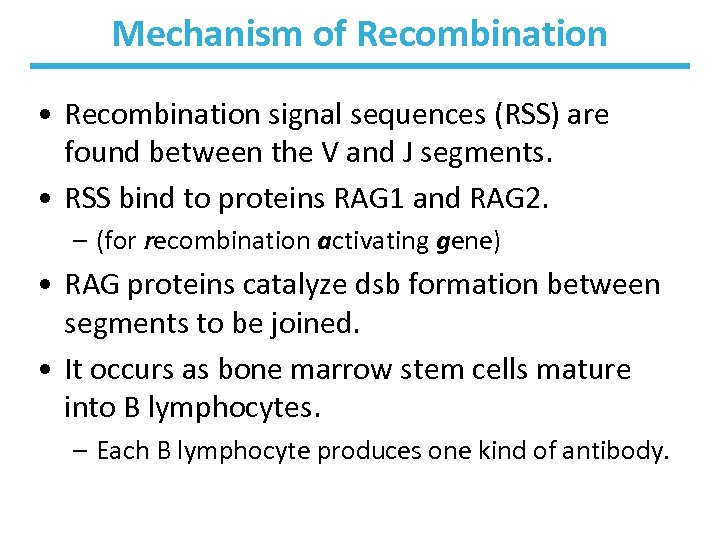
Mechanism of Recombination • Recombination signal sequences (RSS) are found between the V and J segments. • RSS bind to proteins RAG 1 and RAG 2. – (for recombination activating gene) • RAG proteins catalyze dsb formation between segments to be joined. • It occurs as bone marrow stem cells mature into B lymphocytes. – Each B lymphocyte produces one kind of antibody.
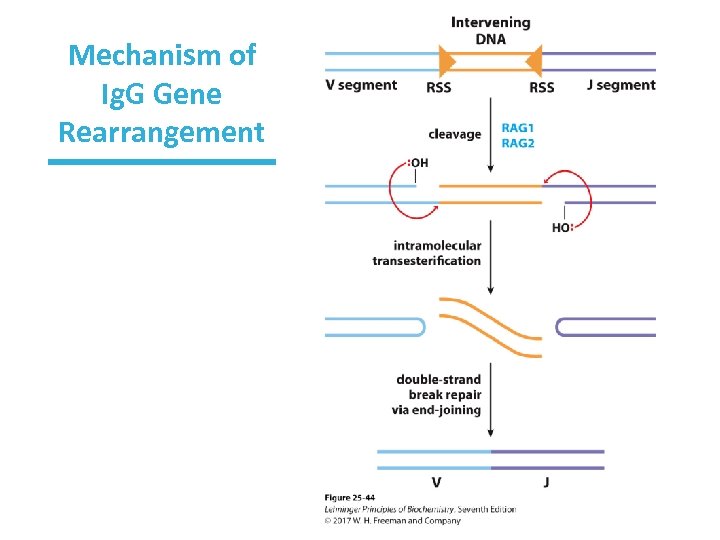
Mechanism of Ig. G Gene Rearrangement

Chapter 25: Summary In this chapter, we learned that: • DNA replication is catalyzed by DNA polymerases that use one of the strands as a template while adding new nucleotides to the 3’-end of the newly synthesized chain • several mechanisms exist to correct mismatches and other changes in DNA • during DNA repair, the information encoded in the parent strand can be used to make corrections in the daughter strand • homologous recombination involves swapping of regions of DNA with similar sequences
2859d3cdc2adf736595518d302f4a70c.ppt