3d77668cbe61c80f5ed8ac6ab1ec68b7.ppt
- Количество слайдов: 46

2 Solid-phase Synthesis and Probe Labeling Processes Purification and Diafiltration Processes for Nucleic Acid Diagnostic Testing (NAT) Products Vũ Mạnh Huỳnh Tiến Sĩ Hóa Học
Solid-phase Synthesis & Labeling Processes for Nucleic Acid Diagnostic Testings (NAT) Phophoramidite chemistry for oligo synthesis: 1. Potential for automated synthesis. 2. High coupling efficiency: 99. 8 – 99. 9% 3. Ease of site-specific: – Non-nucleosidal base. – Linker incorporation. 4. Ready scalability: – AKTA Oligo. Pilot 10/100: 1 - 50µmole (1 gram of 20 mer), 50µmole - 9 mmole (15 grams of 20 mer). – Oligo. Pilot 400: 4 - 30 mmole Scale (150 grams of 20 mer). – Oligo. Process: 50 mmole-1 mole (5 kilograms of 20 mer).

DNA/RNA Synthesizers from GE Healthcare Biosciences
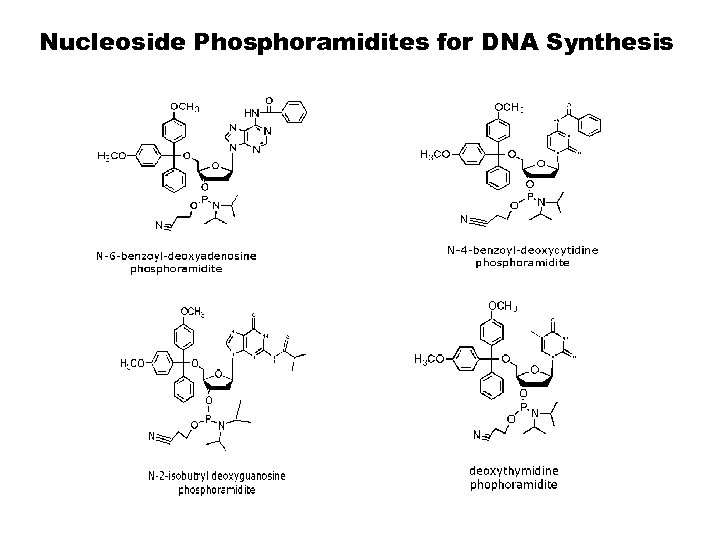
Nucleoside Phosphoramidites for DNA Synthesis
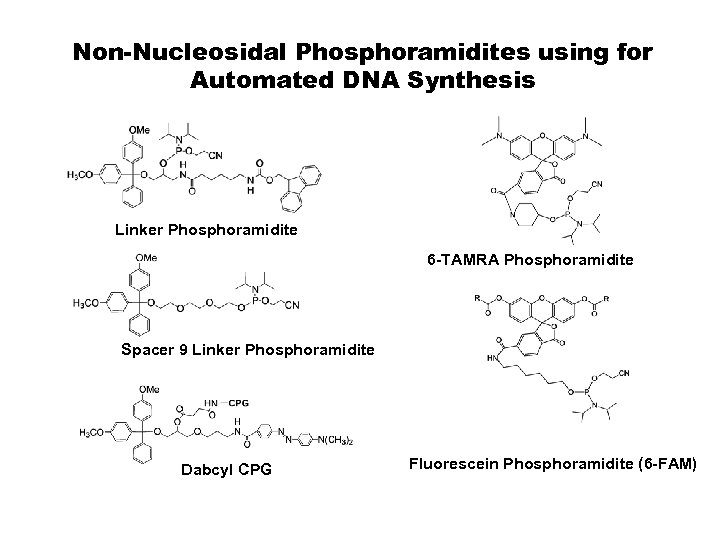
Non-Nucleosidal Phosphoramidites using for Automated DNA Synthesis Linker Phosphoramidite 6 -TAMRA Phosphoramidite Spacer 9 Linker Phosphoramidite Dabcyl CPG Fluorescein Phosphoramidite (6 -FAM)

Automated Solid-Phased Synthesis 1. Oligonucleotides can be rapidly assembled by repeatedly addition of monomers using solid-phase methods provided coupling efficiencies are consistently high. 2. Eliminate isolation & purification after each cycle 3. Syntheses are continuing with four synthesis steps: • Step 1: Detritylation • Step 2: Base Condensation • Step 3: Oxidation • Step 4: Capping Post Synthesis: Cleavage/Deprotection 1. Standard Condition: Conc. Ammonium Hydroxide, at 56 - 60° C, 16 hours. 2. Mild Condition: t-Butylamine : Me-OH : H 2 O; 1: 1: 2, at 56 - 60° C, 16 hours.
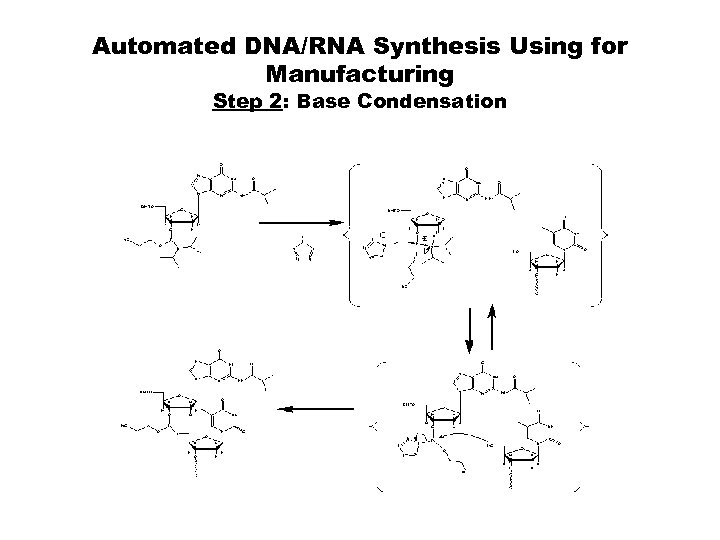
Automated DNA/RNA Synthesis Using for Manufacturing Step 2: Base Condensation
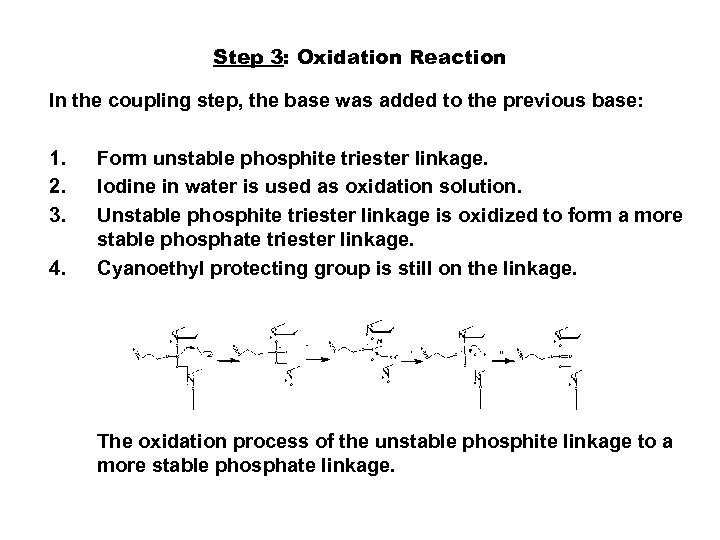
Step 3: Oxidation Reaction In the coupling step, the base was added to the previous base: 1. 2. 3. 4. Form unstable phosphite triester linkage. Iodine in water is used as oxidation solution. Unstable phosphite triester linkage is oxidized to form a more stable phosphate triester linkage. Cyanoethyl protecting group is still on the linkage. The oxidation process of the unstable phosphite linkage to a more stable phosphate linkage.
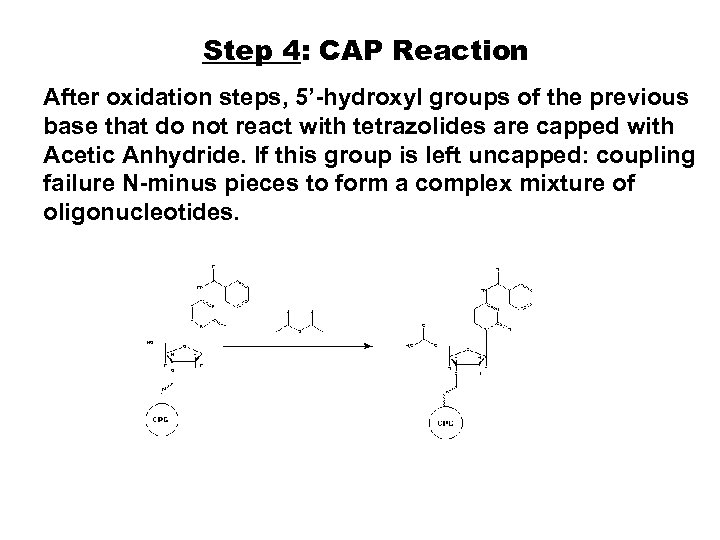
Step 4: CAP Reaction After oxidation steps, 5’-hydroxyl groups of the previous base that do not react with tetrazolides are capped with Acetic Anhydride. If this group is left uncapped: coupling failure N-minus pieces to form a complex mixture of oligonucleotides.
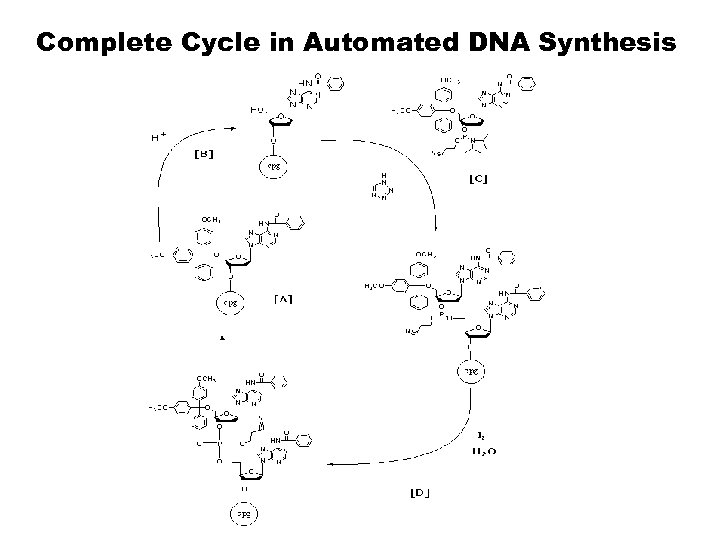
Complete Cycle in Automated DNA Synthesis
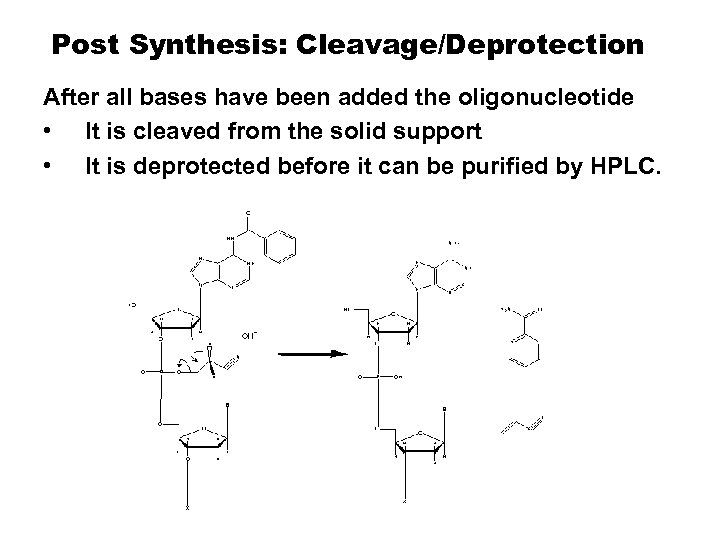
Post Synthesis: Cleavage/Deprotection After all bases have been added the oligonucleotide • It is cleaved from the solid support • It is deprotected before it can be purified by HPLC.
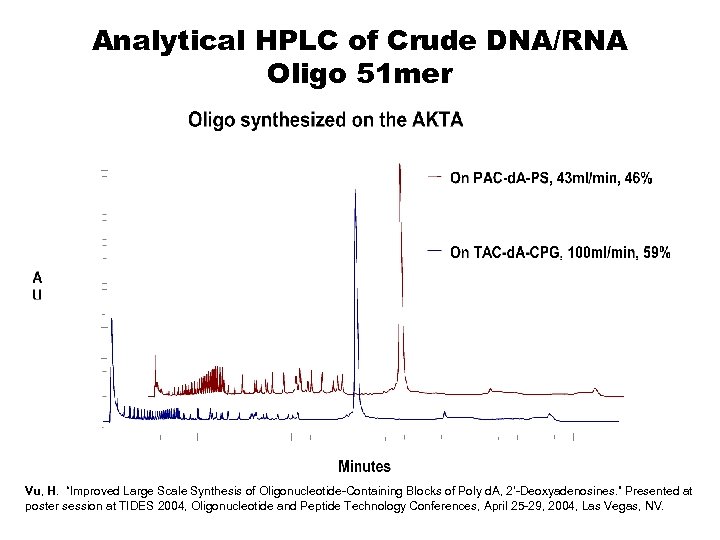
Analytical HPLC of Crude DNA/RNA Oligo 51 mer Vu, H. “Improved Large Scale Synthesis of Oligonucleotide-Containing Blocks of Poly d. A, 2’-Deoxyadenosines. ” Presented at poster session at TIDES 2004, Oligonucleotide and Peptide Technology Conferences, April 25 -29, 2004, Las Vegas, NV.

Manufacturing of Oligonucleotide containing linker After deprotection, crude oligo is collected by filtration to remove solid support. 1. Desired oligo 2. N-Plus pieces 3. N-Minus pieces 4. Protecting groups The crude is purified by RP-HPLC or IEX-HPLC. The pure oligo is subjected to the next step.

Molecular Beacon Torch Manufacturing 1. 2. 3. 4. 5. 6. 7. 8. 9. Labeling process uses modified cycle. Use fresh dye-reagent. 4 minutes coupling. No CAP Step. Deprotect under mild condition. Purify by IEX-HPLC. Unstable in Na. OH. Stable in Sodium Chloride. Process can be scaled up to MFG full-scale.
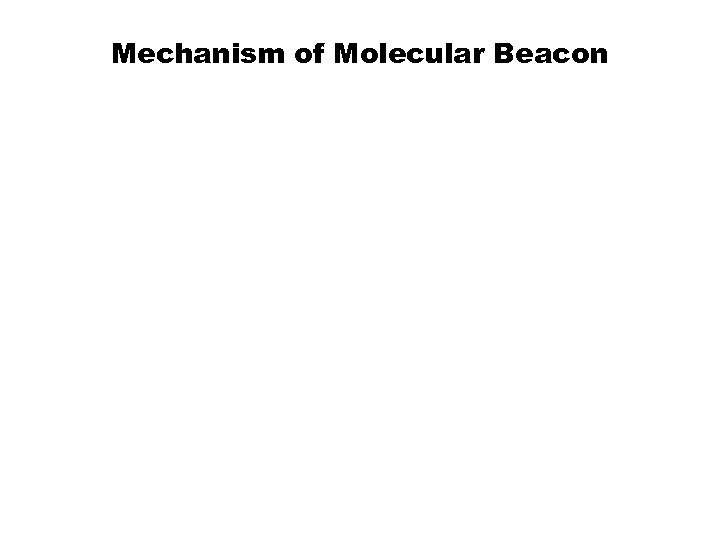
Mechanism of Molecular Beacon

Dyes using in probe synthesis Dyes Ex (nm) Em (nm) Company 1. FAM 494 518 Molecular Probes* 2. Oregon Green 488 496 524 Molecular Probes* 3. Oregon Green 514 511 530 Molecular Probes* 4. Rhodamine Green 502 527 Molecular Probes* 5. 6 -JOE 520 548 Molecular Probes* 6. Carboxyrhodamine 6 G 525 555 Molecular Probes* 7. 6 -TET 521 536 Molecular Probes* 8. HEX 535 556 Molecular Probes* 9. TAMRA 555 580 Molecular Probes* 10. Lissamine Rhodamine B 570 590 Molecular Probes* 11. Rhodamine Red-X 580 590 Molecular Probes* 12. ROX 580 605 Molecular Probes* 13. Texas Red 595 615 Molecular Probes* 14. Naphthofluorescein 602 672 Molecular Probes* 15. BODIPY Dyes var Molecular Probes* 16. Alexa Fluor Dyes var Molecular Probes 17. Cy. Dyes var Amersham Biosciences 18. Oyster Dyes var Denovo Biolabels 19. IRDyes var LI-COR 20. Epoch Pharm dyes var Epoch Pharmaceuticals *Dye available from this company; not necessarily the patent owner

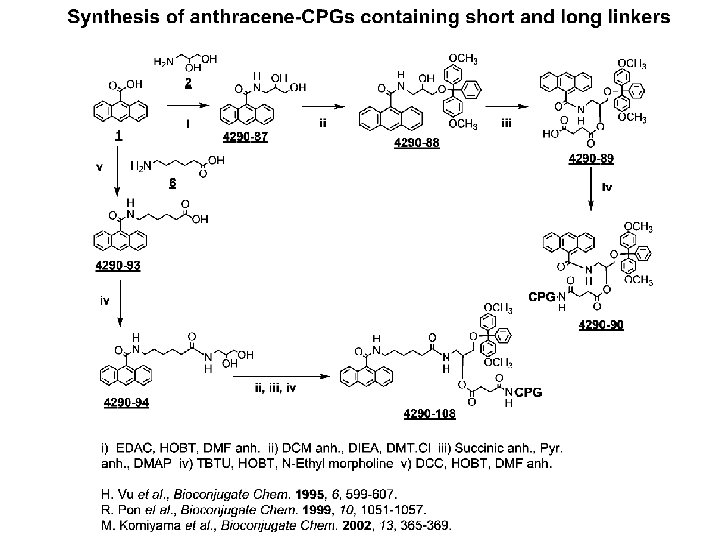
Design & Synthesis of Quencher 1. Cyclic structure, that has molecular model on a plane, such as Benzene ring. This factor will allow quencher to be in a close proximity to the dye. 2. At least one six-member ring. 3. The wave-length, which is closed to that of the dye. 4. Diazo linkages, quinones, carbonyl group. . , those type of compounds would emit higher wave-length. 5. Pyrene (Pyrene-CPG support is commercially available for other purpose in oligonucleotide chemistry). λEx 345 ; λEm 376 6. Acridine (Acridine-CPG support is commercially available as intercalating agent in oligonucleotide chemistry). λEx 480 to 490 7. Napthalene 8. 1, 8 -Naphalic anhydride, 9. Anthracene 10. Xanthene 11. Fluorene 12. Gallocyanine, λEx 610 nm 13. Coumarine, λEx 450 ; λEm 505 nm
Reverse-Phased and Ion Exchanged HPLC Purification Processes Using for MFG. On-Column Detritylation 1. Crude oligos with DMT group, is firstly separated from Non-DMT. 2. N-minus pieces, eluted from the column. 3. Oligo with DMT is detritylated by trifluoro acetic acid (TFA) in the column(OCD). 4. the “DMT-OFF” mixture, is then purified by the same HPLC gradient system.
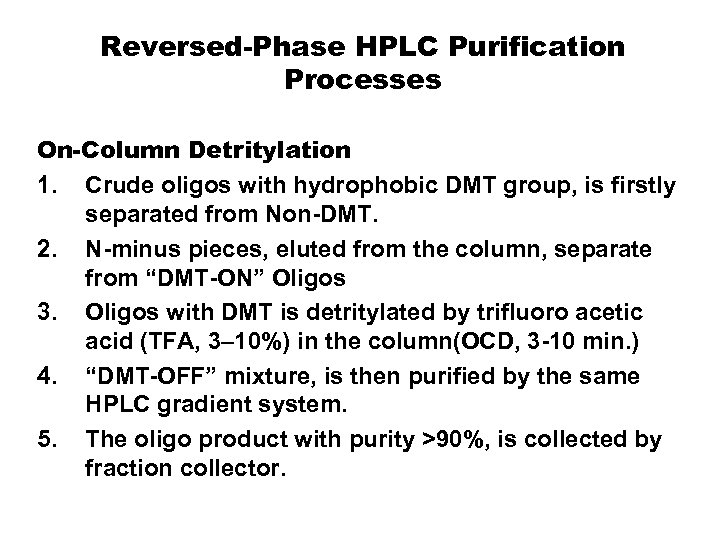
Reversed-Phase HPLC Purification Processes On-Column Detritylation 1. Crude oligos with hydrophobic DMT group, is firstly separated from Non-DMT. 2. N-minus pieces, eluted from the column, separate from “DMT-ON” Oligos 3. Oligos with DMT is detritylated by trifluoro acetic acid (TFA, 3– 10%) in the column(OCD, 3 -10 min. ) 4. “DMT-OFF” mixture, is then purified by the same HPLC gradient system. 5. The oligo product with purity >90%, is collected by fraction collector.

Reversed-Phase Purification Method
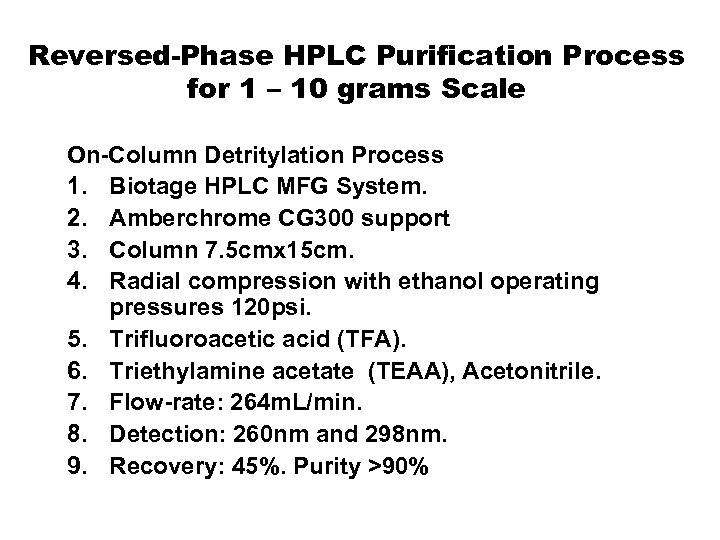
Reversed-Phase HPLC Purification Process for 1 – 10 grams Scale On-Column Detritylation Process 1. Biotage HPLC MFG System. 2. Amberchrome CG 300 support 3. Column 7. 5 cmx 15 cm. 4. Radial compression with ethanol operating pressures 120 psi. 5. Trifluoroacetic acid (TFA). 6. Triethylamine acetate (TEAA), Acetonitrile. 7. Flow-rate: 264 m. L/min. 8. Detection: 260 nm and 298 nm. 9. Recovery: 45%. Purity >90%

Ion Exchange Purification Method
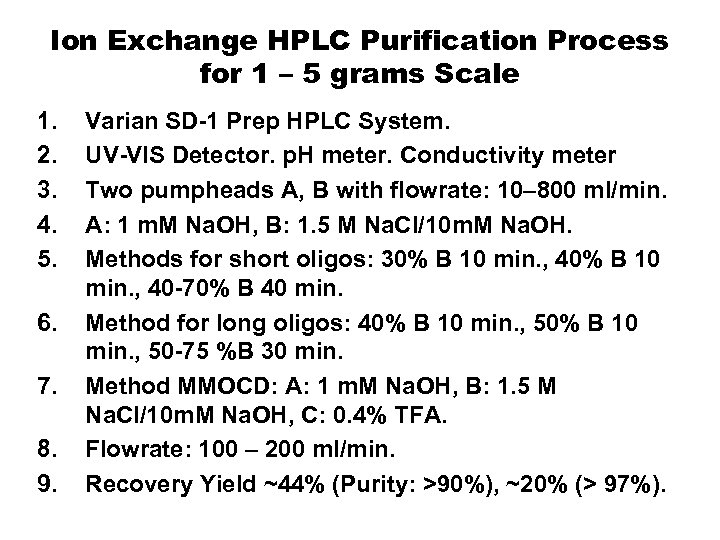
Ion Exchange HPLC Purification Process for 1 – 5 grams Scale 1. 2. 3. 4. 5. 6. 7. 8. 9. Varian SD-1 Prep HPLC System. UV-VIS Detector. p. H meter. Conductivity meter Two pumpheads A, B with flowrate: 10– 800 ml/min. A: 1 m. M Na. OH, B: 1. 5 M Na. Cl/10 m. M Na. OH. Methods for short oligos: 30% B 10 min. , 40 -70% B 40 min. Method for long oligos: 40% B 10 min. , 50 -75 %B 30 min. Method MMOCD: A: 1 m. M Na. OH, B: 1. 5 M Na. Cl/10 m. M Na. OH, C: 0. 4% TFA. Flowrate: 100 – 200 ml/min. Recovery Yield ~44% (Purity: >90%), ~20% (> 97%).
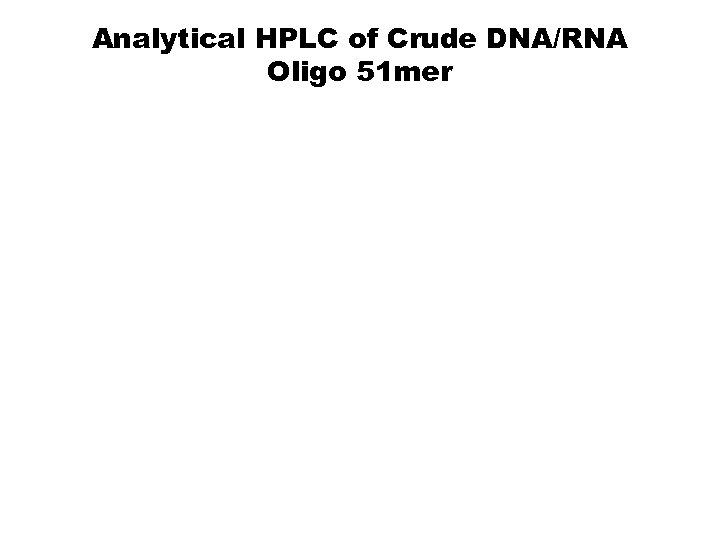
Analytical HPLC of Crude DNA/RNA Oligo 51 mer
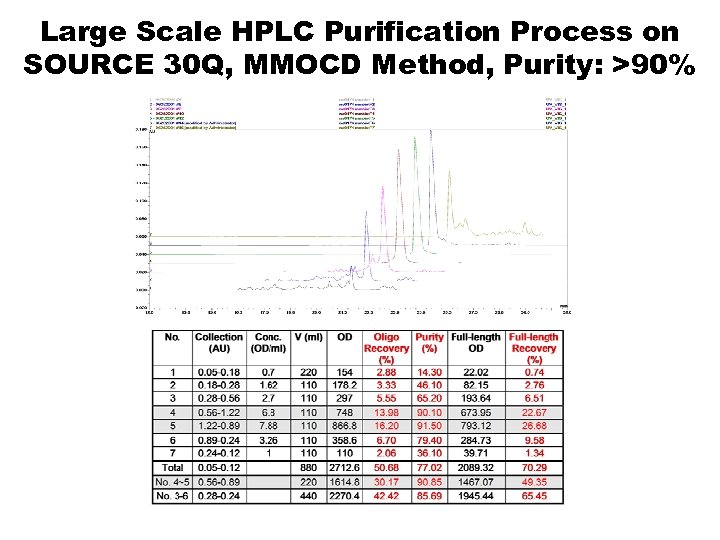
Large Scale HPLC Purification Process on SOURCE 30 Q, MMOCD Method, Purity: >90%
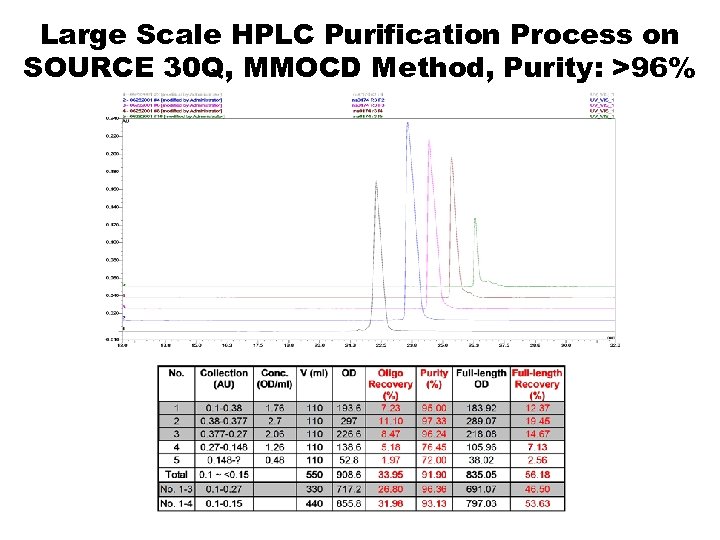
Large Scale HPLC Purification Process on SOURCE 30 Q, MMOCD Method, Purity: >96%
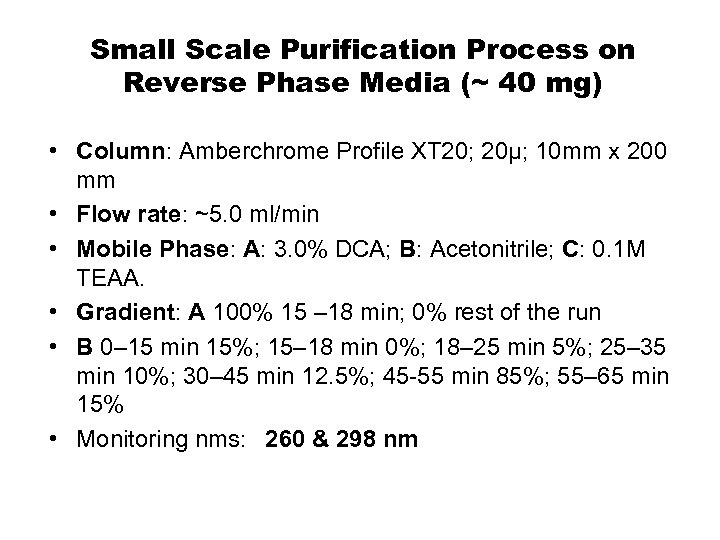
Small Scale Purification Process on Reverse Phase Media (~ 40 mg) • Column: Amberchrome Profile XT 20; 20µ; 10 mm x 200 mm • Flow rate: ~5. 0 ml/min • Mobile Phase: A: 3. 0% DCA; B: Acetonitrile; C: 0. 1 M TEAA. • Gradient: A 100% 15 – 18 min; 0% rest of the run • B 0– 15 min 15%; 15– 18 min 0%; 18– 25 min 5%; 25– 35 min 10%; 30– 45 min 12. 5%; 45 -55 min 85%; 55– 65 min 15% • Monitoring nms: 260 & 298 nm
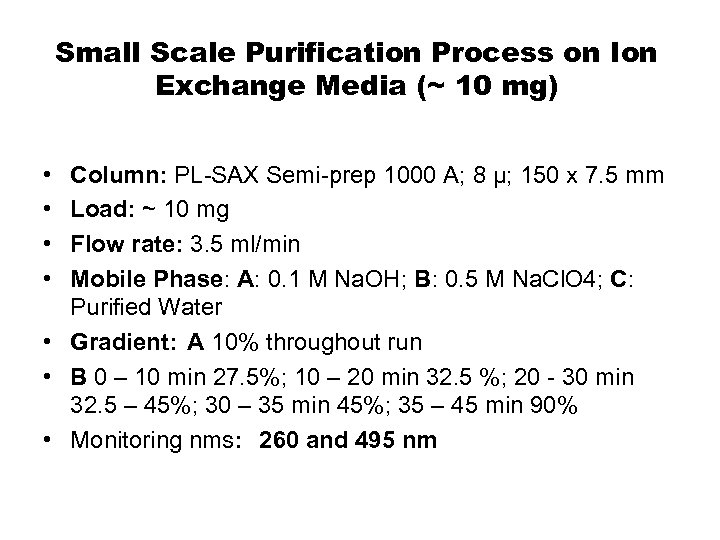
Small Scale Purification Process on Ion Exchange Media (~ 10 mg) • • Column: PL-SAX Semi-prep 1000 A; 8 µ; 150 x 7. 5 mm Load: ~ 10 mg Flow rate: 3. 5 ml/min Mobile Phase: A: 0. 1 M Na. OH; B: 0. 5 M Na. Cl. O 4; C: Purified Water • Gradient: A 10% throughout run • B 0 – 10 min 27. 5%; 10 – 20 min 32. 5 %; 20 - 30 min 32. 5 – 45%; 30 – 35 min 45%; 35 – 45 min 90% • Monitoring nms: 260 and 495 nm

Diafiltration/Desalting Process 1. 2. Ethanol Precipitation using Sodium Acetate. Ultra-filtration: Membrane with MW Cut-Off • A polymer coated membrane • Sodium salt passes through Membrane • Scale: ~ 0. 5 liter per membrane • Problems: Pinholes, Defects 3. Ultra-filtration: Hollow Fiber Cartridges • Similar to Straws • Sodium salt passes through • Avoid problem of membrane • Scale-up to 10 liters • Faster & cheaper • Concentrate to 40 ml
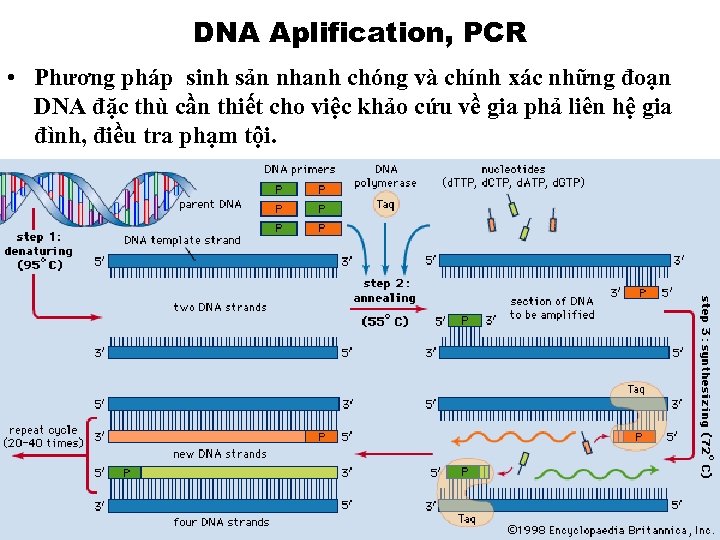
DNA Aplification, PCR • Phương pháp sinh sản nhanh chóng và chính xác những đoạn DNA đặc thù cần thiết cho việc khảo cứu về gia phả liên hệ gia đình, điều tra phạm tội.

In Vitro DNA, RNA Amplification (Phương Pháp PCR, TMA) • Phương pháp amplification được dùng để xác nhận sự tồn tại của virus, tế bào vi sinh vật, động vật, và thực vật • Phương pháp này có thể sản xuất một triệu - một tỷ copies cua di truyền • Amplification đã được đánh giá cao trong khoa học hiện đại, vì phản ứng giữa enzyme và DNA/RNA có thể sinh sản nhanh chóng những DNA rất đặc thù , và được xác nhận dễ dàng bởi hệ thống máy vi tinh. • Một công trình trong vòng 30 phút đến 1 tiếng sẽ cung cấp đầy đủ kết quả chính xác để đi đến kết luận • Vì đặc điểm trên, Amplification đã trở thành một ngành quan trọng cho : Clinical Medicine, Genetic Desease Diagnostic, Forensic Science, Evolutionary biology

Tinh chế DNA, PCR Amplification 1. 2. 3. 4. Thu thập tế bào: Các tế bào chứa DNA được lấy bằng sự cọ xát nhẹ nhàng những que thử, ở trong miệng, trên gần má. Tinh chế tế bào: Tế bào dính trên que thử được ngâm vào trong dung dịch buffer, dùng máy ly tâm để làm cô đọng các tế bào. Tinh chế DNA: Tế bào được hâm nóng để phá màng tế bào, và DNA được ra khỏi tế bào, và hòa tan vao dung dịch PCR Amplification: Khi ở nhiệt độ cao, DNA sẽ dãn ra, và các primers, mang tín hiệu marker, sẽ bám vào đầu của chuỗi DNA, anneal, nhờ điều kiện Thermal Cycler thay đổi, và nhờ enzyme TAQ Polymerase, các vùng quan trọng trên DNA được sinh sản, và trong 1 giờ có thể sinh ra 1 triệu copies.

Phản ứng của công trình Amplification • • PCR Amplification là công trình dùng polymerase enzyme đặc biệt để sinh sản chuỗi complementary từ chuỗi đối tượng DNA, RNA. Trong dung dịch thử DNA, RNA có a) 4 d. NTP, b) 2 primers và c) target sequence. Phản ứng của công trình có 3 giai đoạn 1. Tăng nhiệt độ lên 95° C, để tách rời hai chuỗi của target double helix DNA. 20 giây. 2. Hạ nhiệt độ xuống 55° C, để primers tìm và bám vào vùng thích hợp của chuỗi đã được tách ra. 20 giây. 3. Polymerase enzyme sẽ sinh sản từ primers ra chuỗi complementary mới ở 72° C. 30 giây. Vòng công trình sản xuất này sẽ diễn ra trong vòng 1 tiếng sẽ sinh sản 1 triệu copies.
Real-Time Transcription. Mediated Amplification (RT-TMA) Detection System Vũ Mạnh Huỳnh Tiến Sĩ Hóa Học
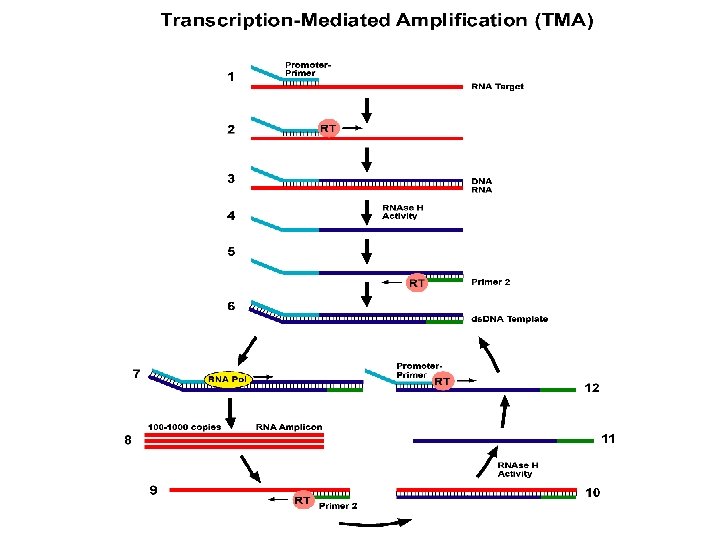
Transcription-Mediated Amplification • • • TMA is an RNA transcription amplification system. Utilizes two enzymes to drive the reaction: 1. T 7 RNA polymerase 2. Reverse transcriptase Amplifies RNA to RNA via DNA intermediates Isothermal reaction, the entire reaction is performed at the same temperature in a water bath or heat block (no thermal-cycler required) Has very rapid kinetics resulting in a billion fold amplification within 15 -30 minutes.
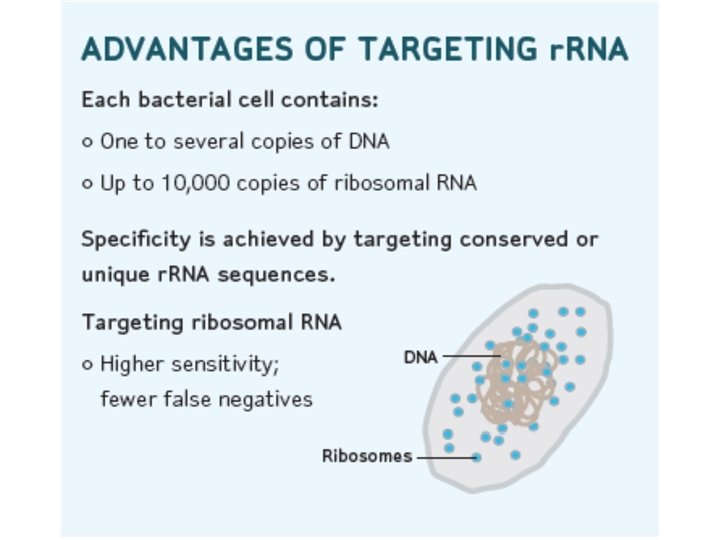
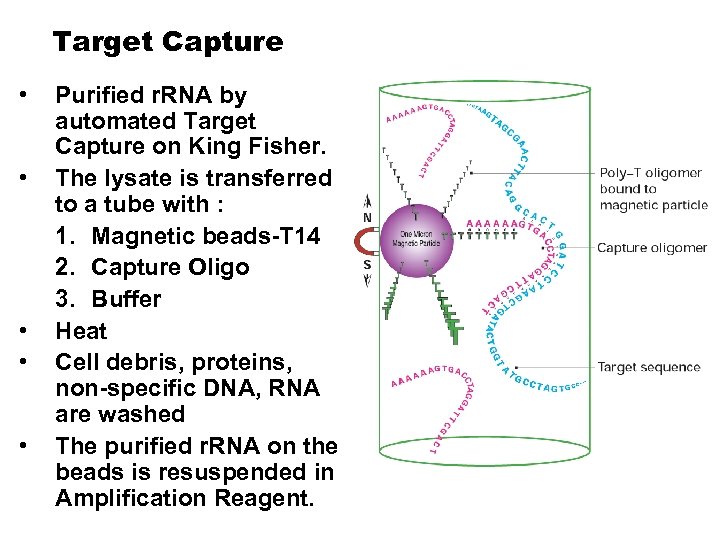
Target Capture • • • Purified r. RNA by automated Target Capture on King Fisher. The lysate is transferred to a tube with : 1. Magnetic beads-T 14 2. Capture Oligo 3. Buffer Heat Cell debris, proteins, non-specific DNA, RNA are washed The purified r. RNA on the beads is resuspended in Amplification Reagent.
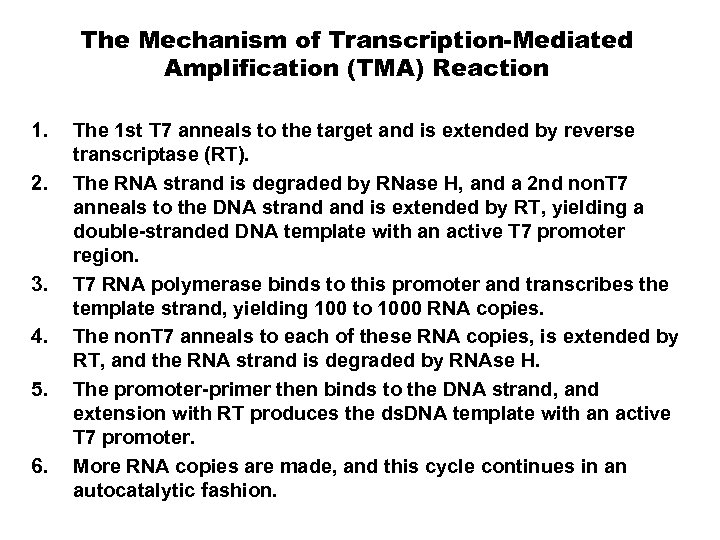
The Mechanism of Transcription-Mediated Amplification (TMA) Reaction 1. 2. 3. 4. 5. 6. The 1 st T 7 anneals to the target and is extended by reverse transcriptase (RT). The RNA strand is degraded by RNase H, and a 2 nd non. T 7 anneals to the DNA strand is extended by RT, yielding a double-stranded DNA template with an active T 7 promoter region. T 7 RNA polymerase binds to this promoter and transcribes the template strand, yielding 100 to 1000 RNA copies. The non. T 7 anneals to each of these RNA copies, is extended by RT, and the RNA strand is degraded by RNAse H. The promoter-primer then binds to the DNA strand, and extension with RT produces the ds. DNA template with an active T 7 promoter. More RNA copies are made, and this cycle continues in an autocatalytic fashion.
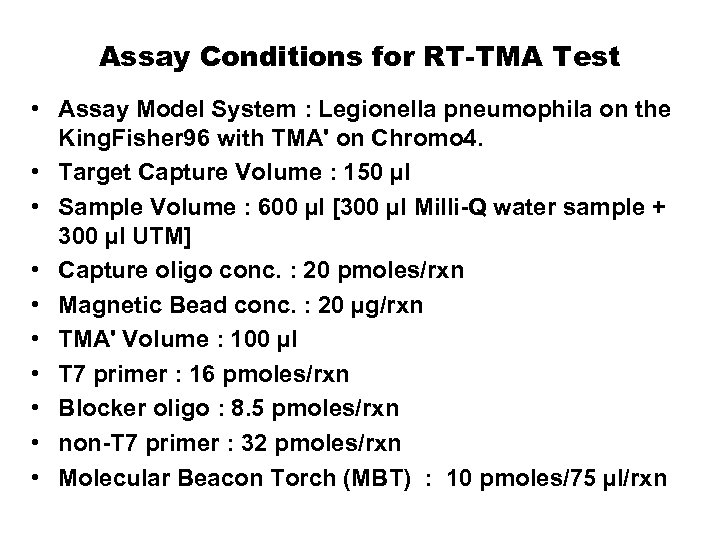
Assay Conditions for RT-TMA Test • Assay Model System : Legionella pneumophila on the King. Fisher 96 with TMA' on Chromo 4. • Target Capture Volume : 150 µl • Sample Volume : 600 µl [300 µl Milli-Q water sample + 300 µl UTM] • Capture oligo conc. : 20 pmoles/rxn • Magnetic Bead conc. : 20 µg/rxn • TMA' Volume : 100 µl • T 7 primer : 16 pmoles/rxn • Blocker oligo : 8. 5 pmoles/rxn • non-T 7 primer : 32 pmoles/rxn • Molecular Beacon Torch (MBT) : 10 pmoles/75 µl/rxn

Real-Time Transcription-Mediated Amplification (RT-TMA) Detection System for Pseudomonas aeruginosa • P. aeruginosa is common organism found in purified water system, or water for injection. • Biofilms from P. aeruginosa can buil up in closed plumbing systems, and equipment. • Contaminations can happen in processes, raw materials, or final product formulations. • The endotoxins of P. aeruginosa can cause serious disease. • It can result in significant quality issues, costly decontamination processes.

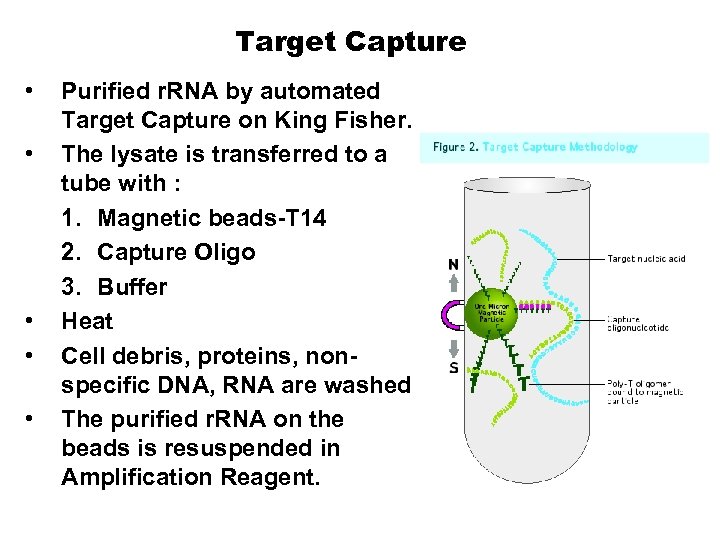
Target Capture • • • Purified r. RNA by automated Target Capture on King Fisher. The lysate is transferred to a tube with : 1. Magnetic beads-T 14 2. Capture Oligo 3. Buffer Heat Cell debris, proteins, nonspecific DNA, RNA are washed The purified r. RNA on the beads is resuspended in Amplification Reagent.
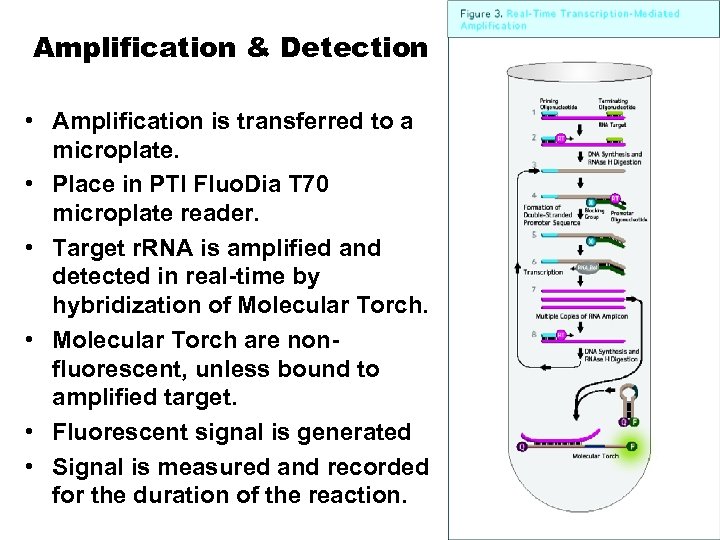
Amplification & Detection • Amplification is transferred to a microplate. • Place in PTI Fluo. Dia T 70 microplate reader. • Target r. RNA is amplified and detected in real-time by hybridization of Molecular Torch. • Molecular Torch are nonfluorescent, unless bound to amplified target. • Fluorescent signal is generated • Signal is measured and recorded for the duration of the reaction.

Real-Time TMA
3d77668cbe61c80f5ed8ac6ab1ec68b7.ppt