386a476e132ac778b151fb3081ae925d.ppt
- Количество слайдов: 42
 1 The PIRSF Protein Classification System as a Basis for Automated Uni. Protein Annotation Icobi 2004 Angra Dos Reis, RJ, Brasil Darren A. Natale, Ph. D. Project Manager and Senior Scientist, PIR Research Assistant Professor, GUMC
1 The PIRSF Protein Classification System as a Basis for Automated Uni. Protein Annotation Icobi 2004 Angra Dos Reis, RJ, Brasil Darren A. Natale, Ph. D. Project Manager and Senior Scientist, PIR Research Assistant Professor, GUMC
 2 Major Topics 1) Uni. Prot Overview 2) PIRSF Protein Classification System 3) Family-Driven Protein Annotation
2 Major Topics 1) Uni. Prot Overview 2) PIRSF Protein Classification System 3) Family-Driven Protein Annotation
 3 Uni. Prot: Universal Protein Resource l l l Central Resource of Protein Sequence and Function International Consortium: PIR, EBI, SIB Unifies PIR-PSD, Swiss-Prot, Tr. EMBL http: //www. uniprot. org
3 Uni. Prot: Universal Protein Resource l l l Central Resource of Protein Sequence and Function International Consortium: PIR, EBI, SIB Unifies PIR-PSD, Swiss-Prot, Tr. EMBL http: //www. uniprot. org
 4 Uni. Prot Databases l l l Uni. Parc: Comprehensive Sequence Archive with Sequence History Uni. Prot: Knowledgebase with Full Classification and Functional Annotation Uni. Ref: Condensed Reference Databases for Sequence Search
4 Uni. Prot Databases l l l Uni. Parc: Comprehensive Sequence Archive with Sequence History Uni. Prot: Knowledgebase with Full Classification and Functional Annotation Uni. Ref: Condensed Reference Databases for Sequence Search
 5 Uni. Parc l An archive for tracking protein sequences l Comprehensive: All published protein sequences Non-Redundant: Merge identical sequence strings Traceable: Versioned, with ‘Active’ or ‘Obsolete’ status tag Concise: no annotation of function, species, tissue, etc. l l l 2. 5 million unique entries from 6 million source-database entries
5 Uni. Parc l An archive for tracking protein sequences l Comprehensive: All published protein sequences Non-Redundant: Merge identical sequence strings Traceable: Versioned, with ‘Active’ or ‘Obsolete’ status tag Concise: no annotation of function, species, tissue, etc. l l l 2. 5 million unique entries from 6 million source-database entries
 6 Uni. Prot Knowledgebase l l Annotated: Fully manually-curated (Swiss-Prot section) and automaticallyannotated based on family-driven rules (Tr. EMBL section) Cross-referenced: Links to over 50 external databases (classification, domain, structure, genome, functional, boutique) Non-redundant: Merge in a single record all protein products derived from a certain gene in a given species High Information Content: l Isoform Presentation: Alternatively Spliced Forms, Proteolytic Cleavage, and Post-Translational Modification (each with FTid) l Nomenclature: Gene/Protein Names (Nomenclature Committees) l Family Classification and Domain Identification: Inter. Pro and PIRSF l Functional Annotation: Function, Functional Site, Developmental Stage, Catalytic Activity, Modification, Regulation, Induction, Pathway, Tissue Specificity, Sub-cellular Location, Disease, Process
6 Uni. Prot Knowledgebase l l Annotated: Fully manually-curated (Swiss-Prot section) and automaticallyannotated based on family-driven rules (Tr. EMBL section) Cross-referenced: Links to over 50 external databases (classification, domain, structure, genome, functional, boutique) Non-redundant: Merge in a single record all protein products derived from a certain gene in a given species High Information Content: l Isoform Presentation: Alternatively Spliced Forms, Proteolytic Cleavage, and Post-Translational Modification (each with FTid) l Nomenclature: Gene/Protein Names (Nomenclature Committees) l Family Classification and Domain Identification: Inter. Pro and PIRSF l Functional Annotation: Function, Functional Site, Developmental Stage, Catalytic Activity, Modification, Regulation, Induction, Pathway, Tissue Specificity, Sub-cellular Location, Disease, Process
 7 Uni. Prot Report • ID & Accession Modified Swiss-Prot “Nice. Prot” view • Name & Taxon • References • Activity • Pathway • Disease • Cross-Refs
7 Uni. Prot Report • ID & Accession Modified Swiss-Prot “Nice. Prot” view • Name & Taxon • References • Activity • Pathway • Disease • Cross-Refs
 8 Uni. Prot Report (II) • Additional Info • Expanded detail Position-specific features: • Active sites • Binding sites • Modified residues • Sequence variations
8 Uni. Prot Report (II) • Additional Info • Expanded detail Position-specific features: • Active sites • Binding sites • Modified residues • Sequence variations
 9 Uni. Ref Databases l Non-Redundant: Merge sequences and subsequences l Uni. Ref 100: 100% sequence identity from all species, including sub-fragments l Superset of Knowledgebase: Includes splice variants and selected Uni. Parc sources (e. g. Ens. EMBL, IPI, and patent data) l Optimized: For Faster Searches using Reduced Data Sets l Uni. Ref 90: 90% sequence identity (36% size reduction) l Uni. Ref 50: 50% sequence identity (63% size reduction)
9 Uni. Ref Databases l Non-Redundant: Merge sequences and subsequences l Uni. Ref 100: 100% sequence identity from all species, including sub-fragments l Superset of Knowledgebase: Includes splice variants and selected Uni. Parc sources (e. g. Ens. EMBL, IPI, and patent data) l Optimized: For Faster Searches using Reduced Data Sets l Uni. Ref 90: 90% sequence identity (36% size reduction) l Uni. Ref 50: 50% sequence identity (63% size reduction)
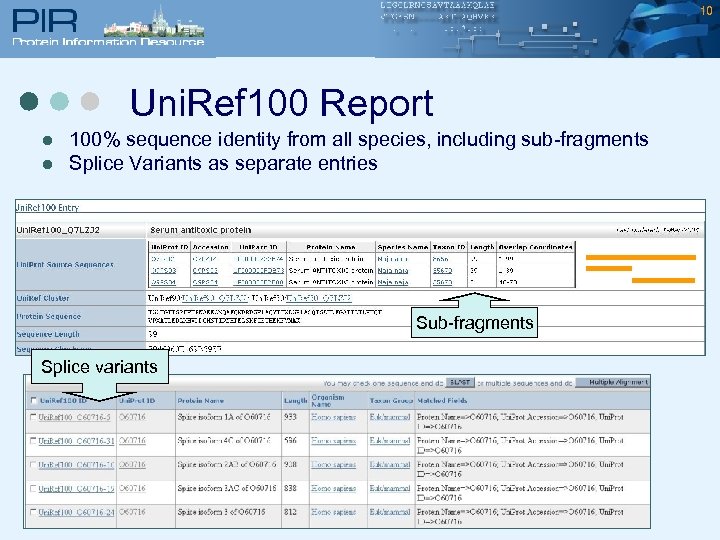 10 Uni. Ref 100 Report l l 100% sequence identity from all species, including sub-fragments Splice Variants as separate entries Sub-fragments Splice variants
10 Uni. Ref 100 Report l l 100% sequence identity from all species, including sub-fragments Splice Variants as separate entries Sub-fragments Splice variants
 11 Uni. Ref 90/50 Reports 90% Phenylalanine hydroxylase sequences likely Merged & have the same function Tryptophan hydroxylase Representative sequence 50%
11 Uni. Ref 90/50 Reports 90% Phenylalanine hydroxylase sequences likely Merged & have the same function Tryptophan hydroxylase Representative sequence 50%
 12 Uni. Prot Web Site http: //www. uniprot. org l Publicly available Dec. 15, 2003 l Text/Sequence Searches against Uni. Prot, Uni. Ref, Uni. Parc l Links to Useful Tools l Download Uni. Prot, Uni. Refs l FAQs and Information l User Help/feedback forms
12 Uni. Prot Web Site http: //www. uniprot. org l Publicly available Dec. 15, 2003 l Text/Sequence Searches against Uni. Prot, Uni. Ref, Uni. Parc l Links to Useful Tools l Download Uni. Prot, Uni. Refs l FAQs and Information l User Help/feedback forms
 13 The Need for Classification Problem: l l l Most new protein sequences come from genome sequencing projects Many have unknown functions Large-scale functional annotation of these sequences based simply on BLAST best hit has pitfalls; results are far from perfect Solution: l Highly curated annotated protein classification system Facilitates: l l Automatic annotation of sequences based on protein families Systematic correction of annotation errors Name standardization in Uni. Prot Functional predictions for uncharacterized proteins This all works only if the system is optimized for annotation
13 The Need for Classification Problem: l l l Most new protein sequences come from genome sequencing projects Many have unknown functions Large-scale functional annotation of these sequences based simply on BLAST best hit has pitfalls; results are far from perfect Solution: l Highly curated annotated protein classification system Facilitates: l l Automatic annotation of sequences based on protein families Systematic correction of annotation errors Name standardization in Uni. Prot Functional predictions for uncharacterized proteins This all works only if the system is optimized for annotation
 14 Levels of Protein Classification Level Example Similarity Evolution Class / Structural elements No relationships Fold TIM-Barrel Topology of backbone Possible monophyly Domain Superfamily Aldolase Recognizable sequence similarity (motifs); basic biochemistry Monophyletic origin Family Class I Aldolase High sequence similarity (alignments); biochemical properties Evolution by ancient duplications Orthologous group 2 -keto-3 -deoxy-6 phosphogluconate aldolase Orthology for a given set of species; biochemical activity; biological function Traceable to a single gene in LCA Lineagespecific expansion (LSE) PA 3131 and PA 3181 Paralogy within a lineage Recent duplication
14 Levels of Protein Classification Level Example Similarity Evolution Class / Structural elements No relationships Fold TIM-Barrel Topology of backbone Possible monophyly Domain Superfamily Aldolase Recognizable sequence similarity (motifs); basic biochemistry Monophyletic origin Family Class I Aldolase High sequence similarity (alignments); biochemical properties Evolution by ancient duplications Orthologous group 2 -keto-3 -deoxy-6 phosphogluconate aldolase Orthology for a given set of species; biochemical activity; biological function Traceable to a single gene in LCA Lineagespecific expansion (LSE) PA 3131 and PA 3181 Paralogy within a lineage Recent duplication
 15 Protein Evolution Sequence changes With enough similarity, one can trace back to a common origin Domain shuffling What about these?
15 Protein Evolution Sequence changes With enough similarity, one can trace back to a common origin Domain shuffling What about these?
 16 Consequences of Domain Shuffling PIRSF 001501 PIRSF 006786 CM (Aro. Q type) PDH CM? CM (Aro. Q type) PDH? PDH CM = chorismate mutase PDH = prephenate dehydrogenase PDT = prephenate dehydratase ACT = regulatory domain PIRSF 001499 PDH PDT? CM/PDH? ACT PIRSF 005547 PDT ACT PIRSF 001424 PDT ACT PIRSF 001500 CM/PDT? CM (Aro. Q type)
16 Consequences of Domain Shuffling PIRSF 001501 PIRSF 006786 CM (Aro. Q type) PDH CM? CM (Aro. Q type) PDH? PDH CM = chorismate mutase PDH = prephenate dehydrogenase PDT = prephenate dehydratase ACT = regulatory domain PIRSF 001499 PDH PDT? CM/PDH? ACT PIRSF 005547 PDT ACT PIRSF 001424 PDT ACT PIRSF 001500 CM/PDT? CM (Aro. Q type)
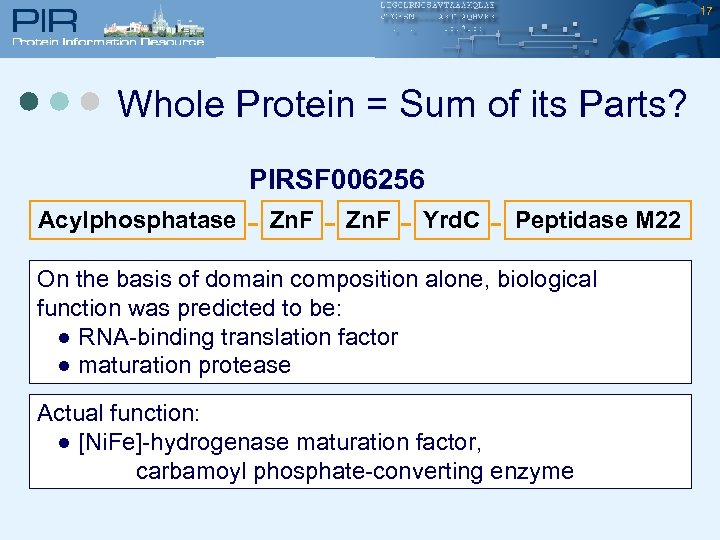 17 Whole Protein = Sum of its Parts? PIRSF 006256 Acylphosphatase - Zn. F - Yrd. C - Peptidase M 22 On the basis of domain composition alone, biological function was predicted to be: ● RNA-binding translation factor ● maturation protease Actual function: ● [Ni. Fe]-hydrogenase maturation factor, carbamoyl phosphate-converting enzyme
17 Whole Protein = Sum of its Parts? PIRSF 006256 Acylphosphatase - Zn. F - Yrd. C - Peptidase M 22 On the basis of domain composition alone, biological function was predicted to be: ● RNA-binding translation factor ● maturation protease Actual function: ● [Ni. Fe]-hydrogenase maturation factor, carbamoyl phosphate-converting enzyme
 18 Classification Goals We strive to reconstruct the natural classification of proteins to the fullest possible extent BUT Domain shuffling rapidly degrades the continuity in the protein structure (faster than sequence divergence degrades similarity) THUS The further we extend the classification, the finer is the domain structure we need to consider SO We need to compromise between the depth of analysis and protein integrity OR Credit: Dr. Y. Wolf, NCBI
18 Classification Goals We strive to reconstruct the natural classification of proteins to the fullest possible extent BUT Domain shuffling rapidly degrades the continuity in the protein structure (faster than sequence divergence degrades similarity) THUS The further we extend the classification, the finer is the domain structure we need to consider SO We need to compromise between the depth of analysis and protein integrity OR Credit: Dr. Y. Wolf, NCBI
 19 Complementary Approaches Whole-protein Classification Domain Classification l l Allows a hierarchy that can trace evolution to the deepest possible level, the last point of traceable homology and common origin Can usually annotate only general biochemical function ØCan l Cannot build a hierarchy deep along the evolutionary tree because of domain shuffling l Can usually annotate specific biological function (preferred to annotate individual proteins) map domains onto proteins classify proteins even when domains are not defined
19 Complementary Approaches Whole-protein Classification Domain Classification l l Allows a hierarchy that can trace evolution to the deepest possible level, the last point of traceable homology and common origin Can usually annotate only general biochemical function ØCan l Cannot build a hierarchy deep along the evolutionary tree because of domain shuffling l Can usually annotate specific biological function (preferred to annotate individual proteins) map domains onto proteins classify proteins even when domains are not defined
 20 The Ideal System… l Comprehensive: each sequence is classified either as a member of a family or as an “orphan” sequence l Hierarchical: families are united into superfamilies on the basis of distant homology, and divided into subfamilies on the basis of close homology l Allows for simultaneous use of the whole protein and domain information (domains mapped onto proteins) l Allows for automatic classification/annotation of new sequences when these sequences are classifiable into the existing families l Expertly curated membership, family name, function, background, etc. l Evidence attribution (experimental vs predicted)
20 The Ideal System… l Comprehensive: each sequence is classified either as a member of a family or as an “orphan” sequence l Hierarchical: families are united into superfamilies on the basis of distant homology, and divided into subfamilies on the basis of close homology l Allows for simultaneous use of the whole protein and domain information (domains mapped onto proteins) l Allows for automatic classification/annotation of new sequences when these sequences are classifiable into the existing families l Expertly curated membership, family name, function, background, etc. l Evidence attribution (experimental vs predicted)
 21 PIRSF Classification System l PIRSF: l l l A network structure from superfamilies to subfamilies Reflects evolutionary relationships of full-length proteins Definitions: l l Homologous: Common ancestry, inferred by sequence similarity l Homeomorphic: Full-length similarity & common domain architecture l l Homeomorphic Family: Basic Unit Network Structure: Flexible number of levels with varying degrees of sequence conservation; allows multiple parents Advantages: l Annotate both general biochemical and specific biological functions l Accurate propagation of annotation and development of standardized protein nomenclature and ontology
21 PIRSF Classification System l PIRSF: l l l A network structure from superfamilies to subfamilies Reflects evolutionary relationships of full-length proteins Definitions: l l Homologous: Common ancestry, inferred by sequence similarity l Homeomorphic: Full-length similarity & common domain architecture l l Homeomorphic Family: Basic Unit Network Structure: Flexible number of levels with varying degrees of sequence conservation; allows multiple parents Advantages: l Annotate both general biochemical and specific biological functions l Accurate propagation of annotation and development of standardized protein nomenclature and ontology
 22 PIRSF Classification System A protein may be assigned to only one homeomorphic family, which may have zero or more child nodes and zero or more parent nodes. Each homeomorphic family may have as many domain superfamily parents as its members have domains.
22 PIRSF Classification System A protein may be assigned to only one homeomorphic family, which may have zero or more child nodes and zero or more parent nodes. Each homeomorphic family may have as many domain superfamily parents as its members have domains.
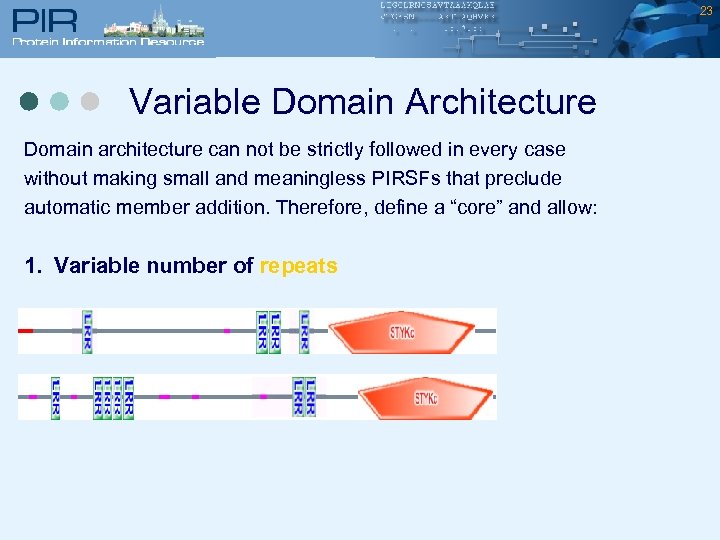 23 Variable Domain Architecture Domain architecture can not be strictly followed in every case without making small and meaningless PIRSFs that preclude automatic member addition. Therefore, define a “core” and allow: 1. Variable number of repeats
23 Variable Domain Architecture Domain architecture can not be strictly followed in every case without making small and meaningless PIRSFs that preclude automatic member addition. Therefore, define a “core” and allow: 1. Variable number of repeats
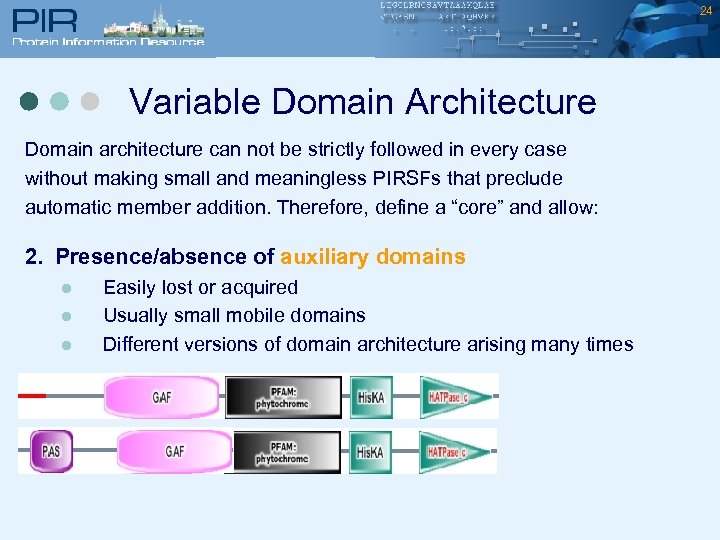 24 Variable Domain Architecture Domain architecture can not be strictly followed in every case without making small and meaningless PIRSFs that preclude automatic member addition. Therefore, define a “core” and allow: 2. Presence/absence of auxiliary domains l l l Easily lost or acquired Usually small mobile domains Different versions of domain architecture arising many times
24 Variable Domain Architecture Domain architecture can not be strictly followed in every case without making small and meaningless PIRSFs that preclude automatic member addition. Therefore, define a “core” and allow: 2. Presence/absence of auxiliary domains l l l Easily lost or acquired Usually small mobile domains Different versions of domain architecture arising many times
 25 Variable Domain Architecture Domain architecture can not be strictly followed in every case without making small and meaningless PIRSFs that preclude automatic member addition. Therefore, define a “core” and allow: 3. Domain duplication
25 Variable Domain Architecture Domain architecture can not be strictly followed in every case without making small and meaningless PIRSFs that preclude automatic member addition. Therefore, define a “core” and allow: 3. Domain duplication
 26 Classification Tool: Blast. Clust l Curator-guided clustering l Retrieve all proteins sharing a common domain l Single-linkage clustering using Blast. Clust l Fixed-length coverage enforces homeomorphicity l Iterative procedure allows tree view
26 Classification Tool: Blast. Clust l Curator-guided clustering l Retrieve all proteins sharing a common domain l Single-linkage clustering using Blast. Clust l Fixed-length coverage enforces homeomorphicity l Iterative procedure allows tree view
 27 PIRSF Family Report (I) Curated family name Description of family Sequence analysis tools Taxonomic distribution of PIRSF can be used to infer evolutionary history of the proteins in the PIRSF Phylogenetic tree and alignment view allows further sequence analysis
27 PIRSF Family Report (I) Curated family name Description of family Sequence analysis tools Taxonomic distribution of PIRSF can be used to infer evolutionary history of the proteins in the PIRSF Phylogenetic tree and alignment view allows further sequence analysis
 28 PIRSF Family Report (II) Integrated value-added information from other databases Mapping to other protein classification databases
28 PIRSF Family Report (II) Integrated value-added information from other databases Mapping to other protein classification databases
 29 Family-Driven Protein Annotation PIRSF Protein Classification provides a platform for Uni. Prot protein annotation l Improve Annotation Quality l Annotate biological function of whole proteins l Annotate uncharacterized hypothetical proteins (functional predictions helped by newly-detected family relationships) Correct annotation errors l Improve under- or over-annotated proteins Standardize Protein Names in Uni. Prot Site annotation l l l
29 Family-Driven Protein Annotation PIRSF Protein Classification provides a platform for Uni. Prot protein annotation l Improve Annotation Quality l Annotate biological function of whole proteins l Annotate uncharacterized hypothetical proteins (functional predictions helped by newly-detected family relationships) Correct annotation errors l Improve under- or over-annotated proteins Standardize Protein Names in Uni. Prot Site annotation l l l
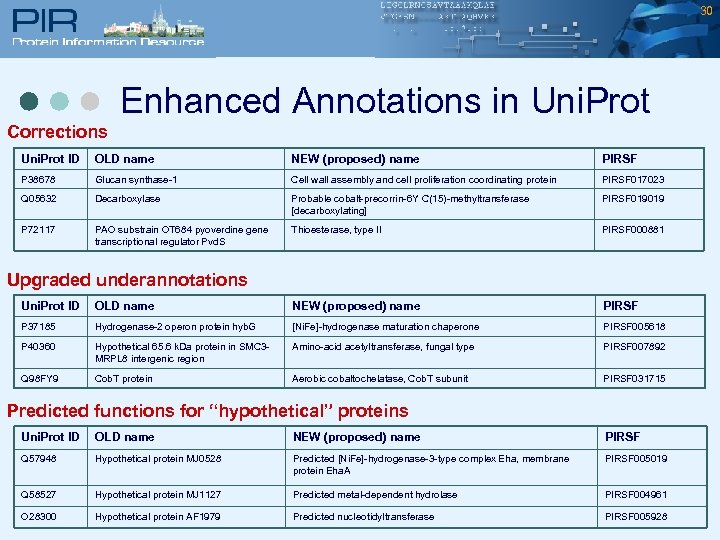 30 Enhanced Annotations in Uni. Prot Corrections Uni. Prot ID OLD name NEW (proposed) name PIRSF P 38678 Glucan synthase-1 Cell wall assembly and cell proliferation coordinating protein PIRSF 017023 Q 05632 Decarboxylase Probable cobalt-precorrin-6 Y C(15)-methyltransferase [decarboxylating] PIRSF 019019 P 72117 PAO substrain OT 684 pyoverdine gene transcriptional regulator Pvd. S Thioesterase, type II PIRSF 000881 Upgraded underannotations Uni. Prot ID OLD name NEW (proposed) name PIRSF P 37185 Hydrogenase-2 operon protein hyb. G [Ni. Fe]-hydrogenase maturation chaperone PIRSF 005618 P 40360 Hypothetical 65. 6 k. Da protein in SMC 3 MRPL 8 intergenic region Amino-acid acetyltransferase, fungal type PIRSF 007892 Q 98 FY 9 Cob. T protein Aerobic cobaltochelatase, Cob. T subunit PIRSF 031715 Predicted functions for “hypothetical” proteins Uni. Prot ID OLD name NEW (proposed) name PIRSF Q 57948 Hypothetical protein MJ 0528 Predicted [Ni. Fe]-hydrogenase-3 -type complex Eha, membrane protein Eha. A PIRSF 005019 Q 58527 Hypothetical protein MJ 1127 Predicted metal-dependent hydrolase PIRSF 004961 O 28300 Hypothetical protein AF 1979 Predicted nucleotidyltransferase PIRSF 005928
30 Enhanced Annotations in Uni. Prot Corrections Uni. Prot ID OLD name NEW (proposed) name PIRSF P 38678 Glucan synthase-1 Cell wall assembly and cell proliferation coordinating protein PIRSF 017023 Q 05632 Decarboxylase Probable cobalt-precorrin-6 Y C(15)-methyltransferase [decarboxylating] PIRSF 019019 P 72117 PAO substrain OT 684 pyoverdine gene transcriptional regulator Pvd. S Thioesterase, type II PIRSF 000881 Upgraded underannotations Uni. Prot ID OLD name NEW (proposed) name PIRSF P 37185 Hydrogenase-2 operon protein hyb. G [Ni. Fe]-hydrogenase maturation chaperone PIRSF 005618 P 40360 Hypothetical 65. 6 k. Da protein in SMC 3 MRPL 8 intergenic region Amino-acid acetyltransferase, fungal type PIRSF 007892 Q 98 FY 9 Cob. T protein Aerobic cobaltochelatase, Cob. T subunit PIRSF 031715 Predicted functions for “hypothetical” proteins Uni. Prot ID OLD name NEW (proposed) name PIRSF Q 57948 Hypothetical protein MJ 0528 Predicted [Ni. Fe]-hydrogenase-3 -type complex Eha, membrane protein Eha. A PIRSF 005019 Q 58527 Hypothetical protein MJ 1127 Predicted metal-dependent hydrolase PIRSF 004961 O 28300 Hypothetical protein AF 1979 Predicted nucleotidyltransferase PIRSF 005928
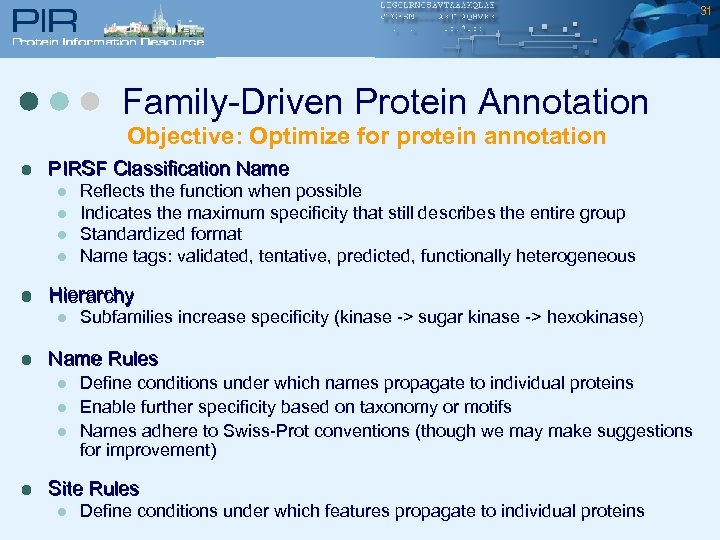 31 Family-Driven Protein Annotation Objective: Optimize for protein annotation l PIRSF Classification Name l l l Hierarchy l l Subfamilies increase specificity (kinase -> sugar kinase -> hexokinase) Name Rules l l Reflects the function when possible Indicates the maximum specificity that still describes the entire group Standardized format Name tags: validated, tentative, predicted, functionally heterogeneous Define conditions under which names propagate to individual proteins Enable further specificity based on taxonomy or motifs Names adhere to Swiss-Prot conventions (though we may make suggestions for improvement) Site Rules l Define conditions under which features propagate to individual proteins
31 Family-Driven Protein Annotation Objective: Optimize for protein annotation l PIRSF Classification Name l l l Hierarchy l l Subfamilies increase specificity (kinase -> sugar kinase -> hexokinase) Name Rules l l Reflects the function when possible Indicates the maximum specificity that still describes the entire group Standardized format Name tags: validated, tentative, predicted, functionally heterogeneous Define conditions under which names propagate to individual proteins Enable further specificity based on taxonomy or motifs Names adhere to Swiss-Prot conventions (though we may make suggestions for improvement) Site Rules l Define conditions under which features propagate to individual proteins
 32 PIR Name Rules l Account for functional variations within one PIRSF, including: l l l Lack of active site residues necessary for enzymatic activity Certain activities relevant only to one part of the taxonomic tree Evolutionarily-related proteins whose biochemical activities are known to differ Monitor such variables to ensure accurate propagation l Propagate other properties that describe function: EC, GO terms, misnomer info, pathway l Name Rule types: l “Zero” Rule l l l Default rule (only condition is membership in the appropriate family) Information is suitable for every member “Higher-Order” Rule l l Has requirements in addition to membership Can have multiple rules that may or may not have mutually exclusive conditions
32 PIR Name Rules l Account for functional variations within one PIRSF, including: l l l Lack of active site residues necessary for enzymatic activity Certain activities relevant only to one part of the taxonomic tree Evolutionarily-related proteins whose biochemical activities are known to differ Monitor such variables to ensure accurate propagation l Propagate other properties that describe function: EC, GO terms, misnomer info, pathway l Name Rule types: l “Zero” Rule l l l Default rule (only condition is membership in the appropriate family) Information is suitable for every member “Higher-Order” Rule l l Has requirements in addition to membership Can have multiple rules that may or may not have mutually exclusive conditions
 33 Example Name Rules Rule ID Rule Conditions Propagated Information PIRNR 00088 1 -1 PIRSF 000881 member and vertebrates Name: S-acyl fatty acid synthase thioesterase EC: oleoyl-[acyl-carrier-protein] hydrolase (EC 3. 1. 2. 14) PIRNR 00088 1 -2 PIRSF 000881 member and not vertebrates Name: Type II thioesterase EC: thiolester hydrolases (EC 3. 1. 2. -) PIRNR 02562 4 -0 PIRSF 025624 member Name: ACT domain protein Misnomer: chorismate mutase Note the lack of a zero rule for PIRSF 000881
33 Example Name Rules Rule ID Rule Conditions Propagated Information PIRNR 00088 1 -1 PIRSF 000881 member and vertebrates Name: S-acyl fatty acid synthase thioesterase EC: oleoyl-[acyl-carrier-protein] hydrolase (EC 3. 1. 2. 14) PIRNR 00088 1 -2 PIRSF 000881 member and not vertebrates Name: Type II thioesterase EC: thiolester hydrolases (EC 3. 1. 2. -) PIRNR 02562 4 -0 PIRSF 025624 member Name: ACT domain protein Misnomer: chorismate mutase Note the lack of a zero rule for PIRSF 000881
 34 Name Rule in Action at Uni. Prot Current: • Automatic annotations (AA) are in a separate field • AA only visible from www. ebi. uniprot. org Future: • Automatic name annotations will become DE line if DE line will improve as a result • AA will be visible from all consortium-hosted web sites
34 Name Rule in Action at Uni. Prot Current: • Automatic annotations (AA) are in a separate field • AA only visible from www. ebi. uniprot. org Future: • Automatic name annotations will become DE line if DE line will improve as a result • AA will be visible from all consortium-hosted web sites
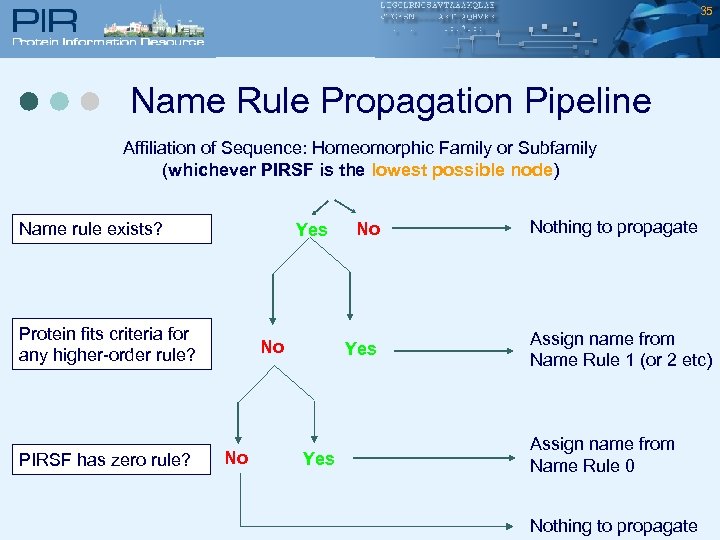 35 Name Rule Propagation Pipeline Affiliation of Sequence: Homeomorphic Family or Subfamily (whichever PIRSF is the lowest possible node) Name rule exists? Yes Protein fits criteria for any higher-order rule? PIRSF has zero rule? No No No Yes Nothing to propagate Assign name from Name Rule 1 (or 2 etc) Assign name from Name Rule 0 Nothing to propagate
35 Name Rule Propagation Pipeline Affiliation of Sequence: Homeomorphic Family or Subfamily (whichever PIRSF is the lowest possible node) Name rule exists? Yes Protein fits criteria for any higher-order rule? PIRSF has zero rule? No No No Yes Nothing to propagate Assign name from Name Rule 1 (or 2 etc) Assign name from Name Rule 0 Nothing to propagate
 36 PIR Site Rules l Position-Specific Site Features: l l Current requirements: l l l active sites binding sites modified amino acids at least one PDB structure experimental data on functional sites: CATRES database (Thornton) Rule Definition: l l Select template structure Align PIRSF seed members with structural template Edit MSA to retain conserved regions covering all site residues Build Site HMM from concatenated conserved regions
36 PIR Site Rules l Position-Specific Site Features: l l Current requirements: l l l active sites binding sites modified amino acids at least one PDB structure experimental data on functional sites: CATRES database (Thornton) Rule Definition: l l Select template structure Align PIRSF seed members with structural template Edit MSA to retain conserved regions covering all site residues Build Site HMM from concatenated conserved regions
 37 Site Rule Algorithm l Match Rule Conditions l Membership Check (PIRSF HMM threshold) l l Ensures that the annotation is appropriate Conserved Region Check (site HMM threshold) Site Residue Check (all position-specific residues in HMMAlign) Propagate Information l l l Feature annotation using controlled vocabulary Evidence attribution (experimental vs. computational prediction) Attribute sources and strengths of evidence
37 Site Rule Algorithm l Match Rule Conditions l Membership Check (PIRSF HMM threshold) l l Ensures that the annotation is appropriate Conserved Region Check (site HMM threshold) Site Residue Check (all position-specific residues in HMMAlign) Propagate Information l l l Feature annotation using controlled vocabulary Evidence attribution (experimental vs. computational prediction) Attribute sources and strengths of evidence
 38 Match Rule Conditions Only propagate site annotation if all rule conditions are met
38 Match Rule Conditions Only propagate site annotation if all rule conditions are met
 39 PIRSF Family Report (III) Defined rules for annotation Site rules allow precise annotation of features for Uni. Prot proteins within the PIRSF
39 PIRSF Family Report (III) Defined rules for annotation Site rules allow precise annotation of features for Uni. Prot proteins within the PIRSF
 40 Site Rules Feed Name Rules Functional variation within one PIRSF: binding sites with different specificity drive choice of applicable rule to ensure appropriate annotation ? Functional Site rule: tags active site, binding, other residue-specific information Functional Annotation rule: gives name, EC, other activity-specific information
40 Site Rules Feed Name Rules Functional variation within one PIRSF: binding sites with different specificity drive choice of applicable rule to ensure appropriate annotation ? Functional Site rule: tags active site, binding, other residue-specific information Functional Annotation rule: gives name, EC, other activity-specific information
 41 PIR Team l l Dr. Cathy Wu, Director Curation team Dr. Winona Barker Dr. Zhangzhi Hu Dr. Raja Mazumder l Dr. Darren Natale Dr. CR Vinayaka Dr. Anastasia Nikolskaya Dr. Xianying Wei Dr. Sona Vasudevan Dr. Lai-Su Yeh Informatics team Dr. Leslie Arminski Yongxing Chen, M. S. Dr. Hsing-Kuo Hua Sehee Chung, M. S. Dr. Hongzhan Huang Baris Suzek, M. S. l Students Jorge Castro-Alvear Christina Fang l Jian Zhang, M. S. Amar Kalelkar Vincent Hormoso Natalia Petrova Uni. Prot Collaborators Dr. Rolf Apweiler/EBI Dr. Amos Bairoch/SIB Rathi Thiagarajan
41 PIR Team l l Dr. Cathy Wu, Director Curation team Dr. Winona Barker Dr. Zhangzhi Hu Dr. Raja Mazumder l Dr. Darren Natale Dr. CR Vinayaka Dr. Anastasia Nikolskaya Dr. Xianying Wei Dr. Sona Vasudevan Dr. Lai-Su Yeh Informatics team Dr. Leslie Arminski Yongxing Chen, M. S. Dr. Hsing-Kuo Hua Sehee Chung, M. S. Dr. Hongzhan Huang Baris Suzek, M. S. l Students Jorge Castro-Alvear Christina Fang l Jian Zhang, M. S. Amar Kalelkar Vincent Hormoso Natalia Petrova Uni. Prot Collaborators Dr. Rolf Apweiler/EBI Dr. Amos Bairoch/SIB Rathi Thiagarajan
 42 Curator’s Decision Maker
42 Curator’s Decision Maker


