bac4863f446c8e79229af4cd0ab9960a.ppt
- Количество слайдов: 40
 1 Gene. TUC: Natural Language Understanding in Medical Text Ph. D Defense Rune Sætre June 27 th 2006
1 Gene. TUC: Natural Language Understanding in Medical Text Ph. D Defense Rune Sætre June 27 th 2006
 2 Overview • Motivation • Thesis Work – – Overview (Diploma Thesis) Idea (Paper 1 and 2) Bioogle (Paper 3, 4 and 5) Gene. TUC (Paper 6) • Results, Related Work and Discussion • Comments and Questions by Jong C. Park and Eivind Hovig
2 Overview • Motivation • Thesis Work – – Overview (Diploma Thesis) Idea (Paper 1 and 2) Bioogle (Paper 3, 4 and 5) Gene. TUC (Paper 6) • Results, Related Work and Discussion • Comments and Questions by Jong C. Park and Eivind Hovig
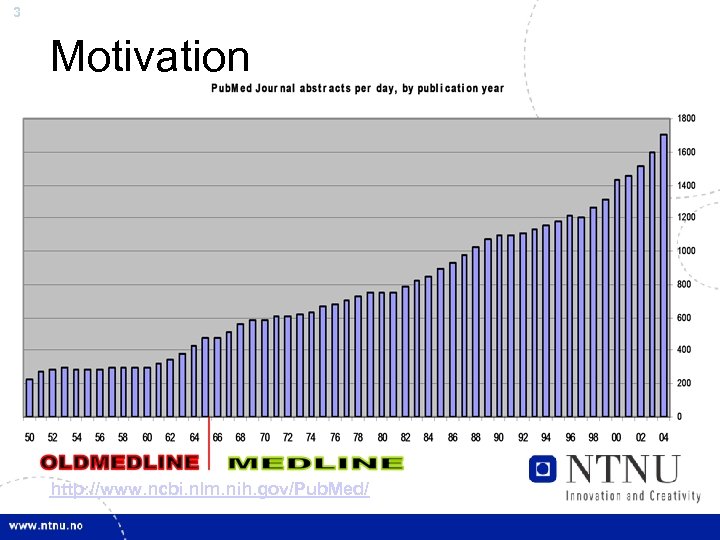 3 Motivation http: //www. ncbi. nlm. nih. gov/Pub. Med/
3 Motivation http: //www. ncbi. nlm. nih. gov/Pub. Med/
 4 …Motivation • Biomedical Researchers publish almost 2000 abstracts per day in MEDLINE – Computers are needed to automatically find all (recall), and only (precision), the relevant information • Future Solution: Gene. TUC – TUC: The Understanding Computer – Bus. TUC works for Natural Language queries about busses in Trondheim – Gene. TUC uses full-parsing to extract knowledge from MEDLINE – After parsing the input, Gene. TUC can answer simple questions about protein and gene interactions and other facts from the text
4 …Motivation • Biomedical Researchers publish almost 2000 abstracts per day in MEDLINE – Computers are needed to automatically find all (recall), and only (precision), the relevant information • Future Solution: Gene. TUC – TUC: The Understanding Computer – Bus. TUC works for Natural Language queries about busses in Trondheim – Gene. TUC uses full-parsing to extract knowledge from MEDLINE – After parsing the input, Gene. TUC can answer simple questions about protein and gene interactions and other facts from the text
 5 Challenge: Medical language • Example Input Sentences: – Subsequently, activated CREB activates transcription of genes essential for proper germ cell differentiation. + – Indeed, Ca 2+/calmodulin binds a complex of RGS 4 and a transition state analog of Galpha i 1 -GDP-Al. F 4 -. * • Medical language is not always natural language: – Complex grammar – Invention of new words/names every day + PMID: 11988318 * Bio. Creative 1 Example, PMID 11988318
5 Challenge: Medical language • Example Input Sentences: – Subsequently, activated CREB activates transcription of genes essential for proper germ cell differentiation. + – Indeed, Ca 2+/calmodulin binds a complex of RGS 4 and a transition state analog of Galpha i 1 -GDP-Al. F 4 -. * • Medical language is not always natural language: – Complex grammar – Invention of new words/names every day + PMID: 11988318 * Bio. Creative 1 Example, PMID 11988318
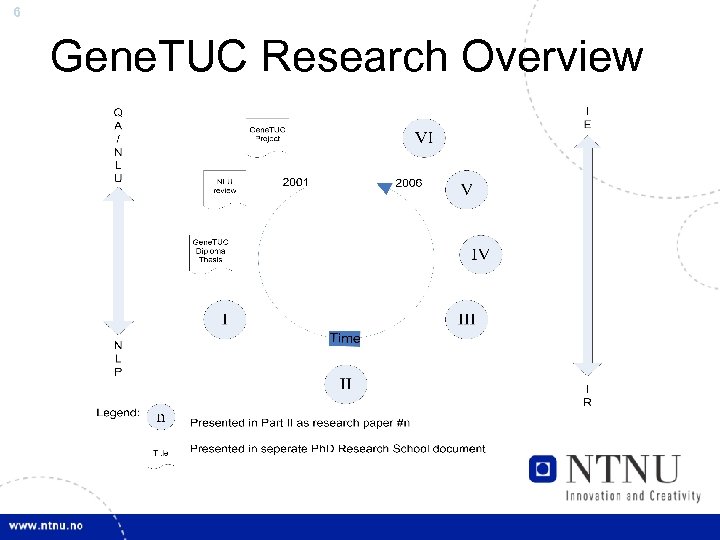 6 Gene. TUC Research Overview
6 Gene. TUC Research Overview
 7 Thesis Work • Gene. TUC Diploma Work – Literature Review: NLU in Medicine – Gene. TUC: Full-parsing of MEDLINE Abstracts • Ph. D Papers: 1 Unitex: Local Grammars 2 Prot. Chew: Automatic Protein Name Recognition 3 Alchymoogle: Automatic Entity Annotation 4 g. Prot: Automatic Protein Interaction Annotation 5 Web. Prot: Online g. Prot Experiments 6 Gene. TUC: GENIA corpus experiments
7 Thesis Work • Gene. TUC Diploma Work – Literature Review: NLU in Medicine – Gene. TUC: Full-parsing of MEDLINE Abstracts • Ph. D Papers: 1 Unitex: Local Grammars 2 Prot. Chew: Automatic Protein Name Recognition 3 Alchymoogle: Automatic Entity Annotation 4 g. Prot: Automatic Protein Interaction Annotation 5 Web. Prot: Online g. Prot Experiments 6 Gene. TUC: GENIA corpus experiments
 8 TUC Introduction • • • Chat-80, Prat-89, HSQL 1991: The Understanding Computer 1996: Bus. TUC (atb. no/bussorakelet/) 2000: Gene. TUC, diploma project 2001 -2006: Gene. TUC has been my Ph. D-Project
8 TUC Introduction • • • Chat-80, Prat-89, HSQL 1991: The Understanding Computer 1996: Bus. TUC (atb. no/bussorakelet/) 2000: Gene. TUC, diploma project 2001 -2006: Gene. TUC has been my Ph. D-Project
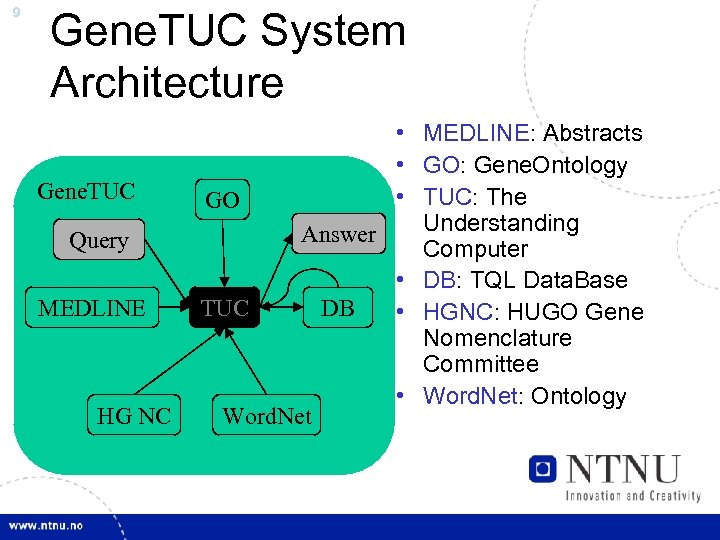 9 Gene. TUC System Architecture Gene. TUC Query MEDLINE HG NC • MEDLINE: Abstracts • GO: Gene. Ontology • TUC: The GO Understanding Answer Computer • DB: TQL Data. Base TUC DB • HGNC: HUGO Gene Nomenclature Committee • Word. Net: Ontology Word. Net
9 Gene. TUC System Architecture Gene. TUC Query MEDLINE HG NC • MEDLINE: Abstracts • GO: Gene. Ontology • TUC: The GO Understanding Answer Computer • DB: TQL Data. Base TUC DB • HGNC: HUGO Gene Nomenclature Committee • Word. Net: Ontology Word. Net
 10 Word. Net 2. 0 • Online lexical reference system – Nouns, verbs, adjectives and adverbs • Inspired by psycholinguistic theories of human lexical memory – Organized into synonym sets, each representing one underlying lexical concept • Different relations link the synonym sets – E. g. hypernyms, hyponyms, holonyms, synonyms, coordinate terms, domain,
10 Word. Net 2. 0 • Online lexical reference system – Nouns, verbs, adjectives and adverbs • Inspired by psycholinguistic theories of human lexical memory – Organized into synonym sets, each representing one underlying lexical concept • Different relations link the synonym sets – E. g. hypernyms, hyponyms, holonyms, synonyms, coordinate terms, domain,
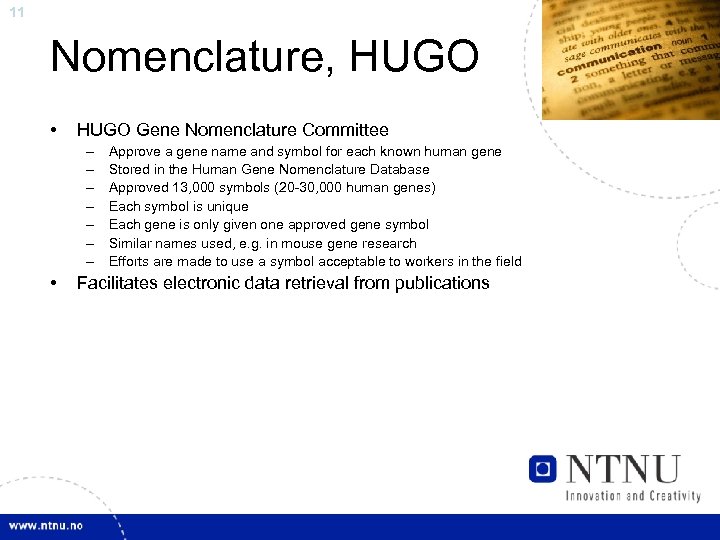 11 Nomenclature, HUGO • HUGO Gene Nomenclature Committee – – – – • Approve a gene name and symbol for each known human gene Stored in the Human Gene Nomenclature Database Approved 13, 000 symbols (20 -30, 000 human genes) Each symbol is unique Each gene is only given one approved gene symbol Similar names used, e. g. in mouse gene research Efforts are made to use a symbol acceptable to workers in the field Facilitates electronic data retrieval from publications
11 Nomenclature, HUGO • HUGO Gene Nomenclature Committee – – – – • Approve a gene name and symbol for each known human gene Stored in the Human Gene Nomenclature Database Approved 13, 000 symbols (20 -30, 000 human genes) Each symbol is unique Each gene is only given one approved gene symbol Similar names used, e. g. in mouse gene research Efforts are made to use a symbol acceptable to workers in the field Facilitates electronic data retrieval from publications
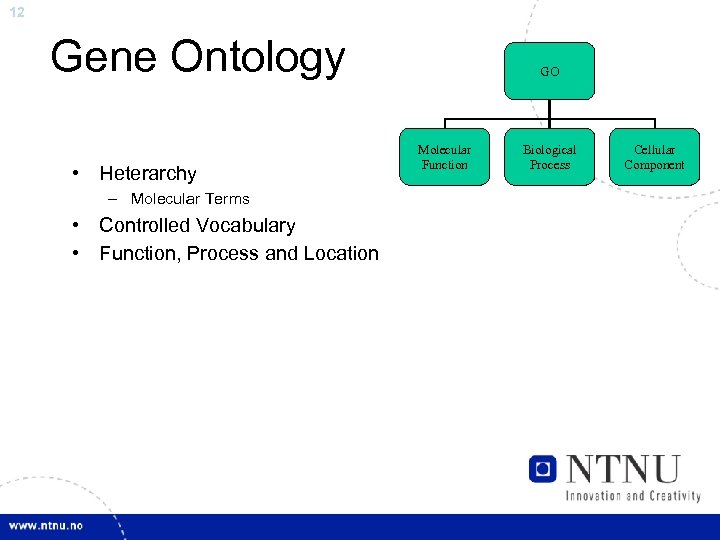 12 Gene Ontology • Heterarchy – Molecular Terms • Controlled Vocabulary • Function, Process and Location GO Molecular Function Biological Process Cellular Component
12 Gene Ontology • Heterarchy – Molecular Terms • Controlled Vocabulary • Function, Process and Location GO Molecular Function Biological Process Cellular Component
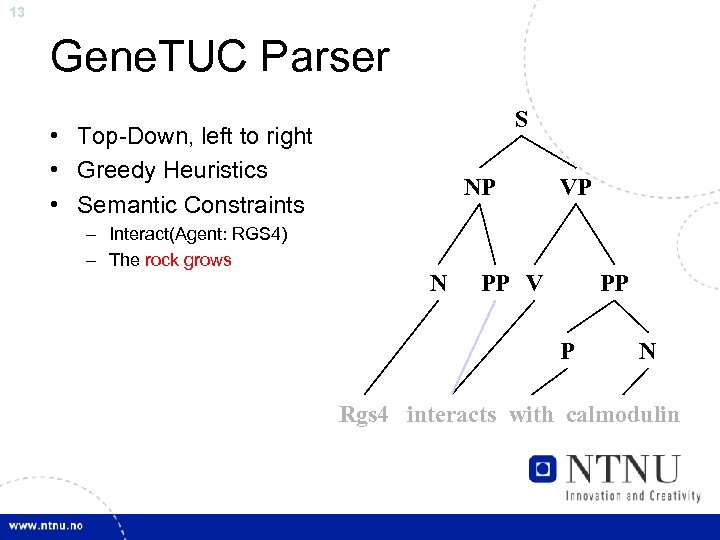 13 Gene. TUC Parser S • Top-Down, left to right • Greedy Heuristics • Semantic Constraints – Interact(Agent: RGS 4) – The rock grows NP N VP PP V PP P N Rgs 4 interacts with calmodulin
13 Gene. TUC Parser S • Top-Down, left to right • Greedy Heuristics • Semantic Constraints – Interact(Agent: RGS 4) – The rock grows NP N VP PP V PP P N Rgs 4 interacts with calmodulin
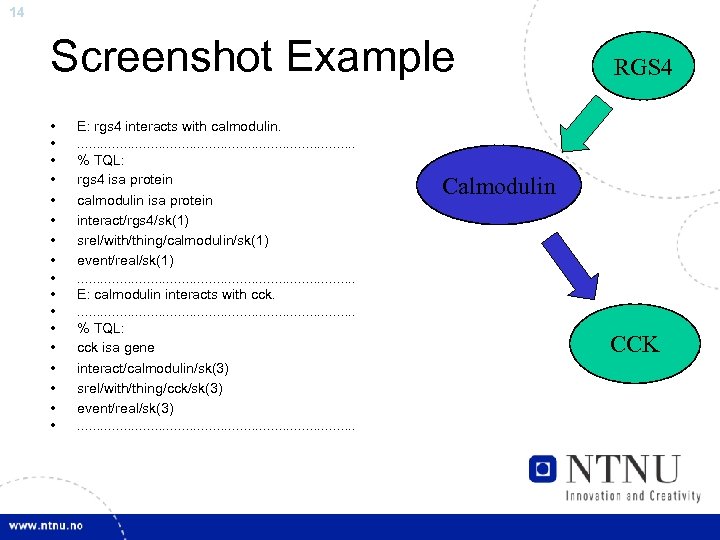 14 Screenshot Example • • • • • E: rgs 4 interacts with calmodulin. . . . . % TQL: rgs 4 isa protein calmodulin isa protein interact/rgs 4/sk(1) srel/with/thing/calmodulin/sk(1) event/real/sk(1). . . . . E: calmodulin interacts with cck. . . . . % TQL: cck isa gene interact/calmodulin/sk(3) srel/with/thing/cck/sk(3) event/real/sk(3). . . . . RGS 4 Calmodulin CCK
14 Screenshot Example • • • • • E: rgs 4 interacts with calmodulin. . . . . % TQL: rgs 4 isa protein calmodulin isa protein interact/rgs 4/sk(1) srel/with/thing/calmodulin/sk(1) event/real/sk(1). . . . . E: calmodulin interacts with cck. . . . . % TQL: cck isa gene interact/calmodulin/sk(3) srel/with/thing/cck/sk(3) event/real/sk(3). . . . . RGS 4 Calmodulin CCK
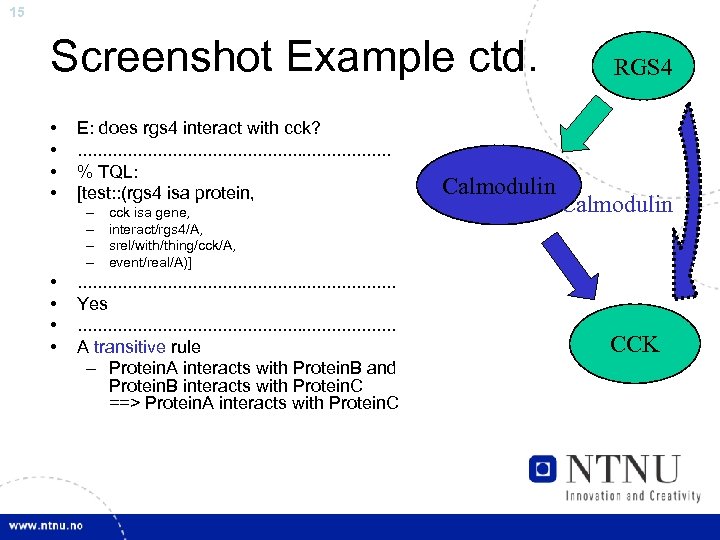 15 Screenshot Example ctd. • • E: does rgs 4 interact with cck? . . . . % TQL: [test: : (rgs 4 isa protein, – – • • cck isa gene, interact/rgs 4/A, srel/with/thing/cck/A, event/real/A)] . . . . Yes. . . . A transitive rule – Protein. A interacts with Protein. B and Protein. B interacts with Protein. C ==> Protein. A interacts with Protein. C Calmodulin RGS 4 Calmodulin CCK
15 Screenshot Example ctd. • • E: does rgs 4 interact with cck? . . . . % TQL: [test: : (rgs 4 isa protein, – – • • cck isa gene, interact/rgs 4/A, srel/with/thing/cck/A, event/real/A)] . . . . Yes. . . . A transitive rule – Protein. A interacts with Protein. B and Protein. B interacts with Protein. C ==> Protein. A interacts with Protein. C Calmodulin RGS 4 Calmodulin CCK
 16 Dictionary • Gene. TUC does not perform very well without a complete dictionary • Current Solution: Bioogle can build a dictionary
16 Dictionary • Gene. TUC does not perform very well without a complete dictionary • Current Solution: Bioogle can build a dictionary
 17 Bioogle (Paper III) • Current ontology: 275 medical terms • Connect Unknowns to these Concepts • Query syntax – “ Unknown is (an|a) “ • Parse results until a hit is found (or not) – “Pentagastrin is a synthetic peptide containing the five terminal amino acids of gastrin. ” • Result: 104 of 200 terms were correctly classified
17 Bioogle (Paper III) • Current ontology: 275 medical terms • Connect Unknowns to these Concepts • Query syntax – “ Unknown is (an|a) “ • Parse results until a hit is found (or not) – “Pentagastrin is a synthetic peptide containing the five terminal amino acids of gastrin. ” • Result: 104 of 200 terms were correctly classified
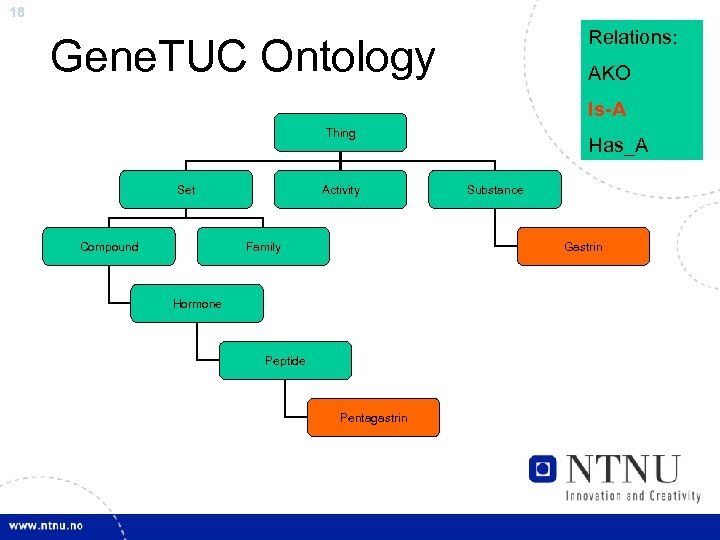 18 Relations: Gene. TUC Ontology AKO Is-A Thing Set Compound Activity Family Has_A Substance Gastrin Hormone Peptide Pentagastrin
18 Relations: Gene. TUC Ontology AKO Is-A Thing Set Compound Activity Family Has_A Substance Gastrin Hormone Peptide Pentagastrin
 19 Google API Search • 1000 queries per user pr day • Free to use for everybody • Can be programmed with SOAP in most languages – Simple Object Access Protocol • Results are handled automatically • Alexa (Amazon) has implemented a similar service* – – $1 per processor hour $1 per gigabyte/year of user storage $1 per 50 gigabytes of data processed $1 per gigabyte uploaded/downloaded * http: //news. bbc. co. uk/1/hi/technology/4530978. stm
19 Google API Search • 1000 queries per user pr day • Free to use for everybody • Can be programmed with SOAP in most languages – Simple Object Access Protocol • Results are handled automatically • Alexa (Amazon) has implemented a similar service* – – $1 per processor hour $1 per gigabyte/year of user storage $1 per 50 gigabytes of data processed $1 per gigabyte uploaded/downloaded * http: //news. bbc. co. uk/1/hi/technology/4530978. stm
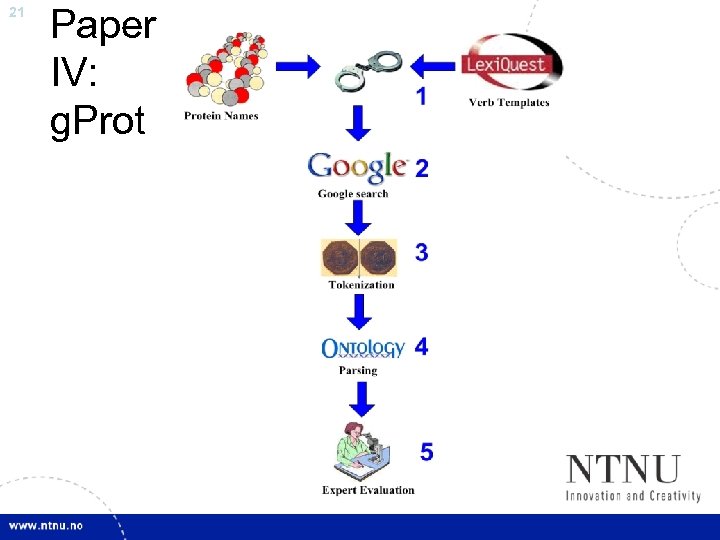
 20 Paper IV: g. Prot • What about protein interactions? • Protein Interaction – Protein – Bio. Cre. At. Iv. E 1: Protein Set of Gene. Ontology Terms • Find publicly known interactions for a given protein, using Google as the main source for new knowledge – Query: “ protein. X Verb. Y “ – Example: “ Gastrin activates “
20 Paper IV: g. Prot • What about protein interactions? • Protein Interaction – Protein – Bio. Cre. At. Iv. E 1: Protein Set of Gene. Ontology Terms • Find publicly known interactions for a given protein, using Google as the main source for new knowledge – Query: “ protein. X Verb. Y “ – Example: “ Gastrin activates “
 21 Paper IV: g. Prot
21 Paper IV: g. Prot
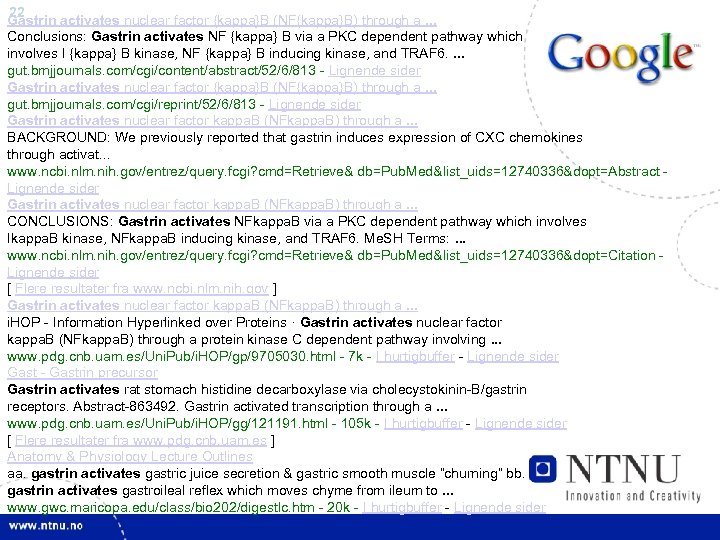 22 Gastrin activates nuclear factor {kappa}B (NF{kappa}B) through a. . . Conclusions: Gastrin activates NF {kappa} B via a PKC dependent pathway which involves I {kappa} B kinase, NF {kappa} B inducing kinase, and TRAF 6. . gut. bmjjournals. com/cgi/content/abstract/52/6/813 - Lignende sider Gastrin activates nuclear factor {kappa}B (NF{kappa}B) through a. . . gut. bmjjournals. com/cgi/reprint/52/6/813 - Lignende sider Gastrin activates nuclear factor kappa. B (NFkappa. B) through a. . . BACKGROUND: We previously reported that gastrin induces expression of CXC chemokines through activat. . . www. ncbi. nlm. nih. gov/entrez/query. fcgi? cmd=Retrieve& db=Pub. Med&list_uids=12740336&dopt=Abstract - Lignende sider Gastrin activates nuclear factor kappa. B (NFkappa. B) through a. . . CONCLUSIONS: Gastrin activates NFkappa. B via a PKC dependent pathway which involves Ikappa. B kinase, NFkappa. B inducing kinase, and TRAF 6. Me. SH Terms: . . . www. ncbi. nlm. nih. gov/entrez/query. fcgi? cmd=Retrieve& db=Pub. Med&list_uids=12740336&dopt=Citation - Lignende sider [ Flere resultater fra www. ncbi. nlm. nih. gov ] Gastrin activates nuclear factor kappa. B (NFkappa. B) through a. . . i. HOP - Information Hyperlinked over Proteins · Gastrin activates nuclear factor kappa. B (NFkappa. B) through a protein kinase C dependent pathway involving. . . www. pdg. cnb. uam. es/Uni. Pub/i. HOP/gp/9705030. html - 7 k - I hurtigbuffer - Lignende sider Gast - Gastrin precursor Gastrin activates rat stomach histidine decarboxylase via cholecystokinin-B/gastrin receptors. Abstract-863492. Gastrin activated transcription through a. . . www. pdg. cnb. uam. es/Uni. Pub/i. HOP/gg/121191. html - 105 k - I hurtigbuffer - Lignende sider [ Flere resultater fra www. pdg. cnb. uam. es ] Anatomy & Physiology Lecture Outlines aa. gastrin activates gastric juice secretion & gastric smooth muscle “churning” bb. gastrin activates gastroileal reflex which moves chyme from ileum to. . . www. gwc. maricopa. edu/class/bio 202/digestlc. htm - 20 k - I hurtigbuffer - Lignende sider
22 Gastrin activates nuclear factor {kappa}B (NF{kappa}B) through a. . . Conclusions: Gastrin activates NF {kappa} B via a PKC dependent pathway which involves I {kappa} B kinase, NF {kappa} B inducing kinase, and TRAF 6. . gut. bmjjournals. com/cgi/content/abstract/52/6/813 - Lignende sider Gastrin activates nuclear factor {kappa}B (NF{kappa}B) through a. . . gut. bmjjournals. com/cgi/reprint/52/6/813 - Lignende sider Gastrin activates nuclear factor kappa. B (NFkappa. B) through a. . . BACKGROUND: We previously reported that gastrin induces expression of CXC chemokines through activat. . . www. ncbi. nlm. nih. gov/entrez/query. fcgi? cmd=Retrieve& db=Pub. Med&list_uids=12740336&dopt=Abstract - Lignende sider Gastrin activates nuclear factor kappa. B (NFkappa. B) through a. . . CONCLUSIONS: Gastrin activates NFkappa. B via a PKC dependent pathway which involves Ikappa. B kinase, NFkappa. B inducing kinase, and TRAF 6. Me. SH Terms: . . . www. ncbi. nlm. nih. gov/entrez/query. fcgi? cmd=Retrieve& db=Pub. Med&list_uids=12740336&dopt=Citation - Lignende sider [ Flere resultater fra www. ncbi. nlm. nih. gov ] Gastrin activates nuclear factor kappa. B (NFkappa. B) through a. . . i. HOP - Information Hyperlinked over Proteins · Gastrin activates nuclear factor kappa. B (NFkappa. B) through a protein kinase C dependent pathway involving. . . www. pdg. cnb. uam. es/Uni. Pub/i. HOP/gp/9705030. html - 7 k - I hurtigbuffer - Lignende sider Gast - Gastrin precursor Gastrin activates rat stomach histidine decarboxylase via cholecystokinin-B/gastrin receptors. Abstract-863492. Gastrin activated transcription through a. . . www. pdg. cnb. uam. es/Uni. Pub/i. HOP/gg/121191. html - 105 k - I hurtigbuffer - Lignende sider [ Flere resultater fra www. pdg. cnb. uam. es ] Anatomy & Physiology Lecture Outlines aa. gastrin activates gastric juice secretion & gastric smooth muscle “churning” bb. gastrin activates gastroileal reflex which moves chyme from ileum to. . . www. gwc. maricopa. edu/class/bio 202/digestlc. htm - 20 k - I hurtigbuffer - Lignende sider
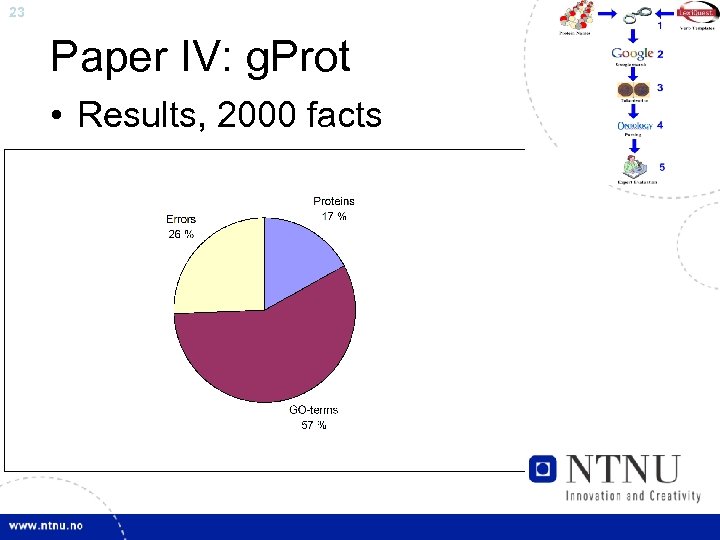 23 Paper IV: g. Prot • Results, 2000 facts
23 Paper IV: g. Prot • Results, 2000 facts
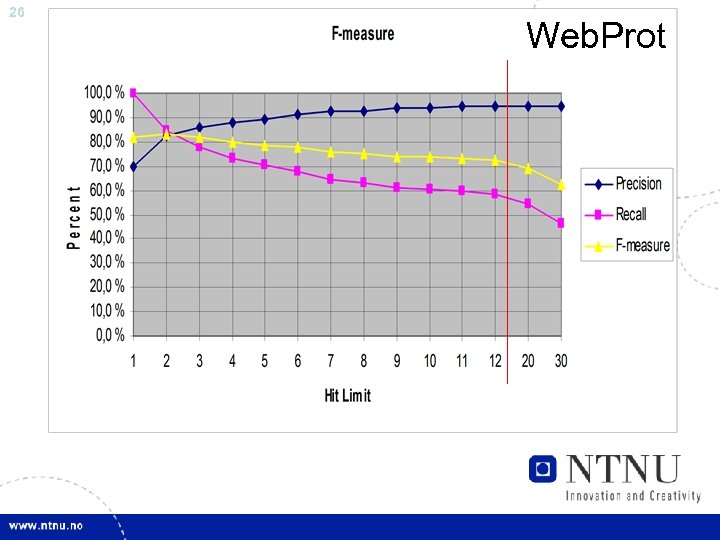
 24 Paper V: Web. Prot • Online Implementation, bigger experiment • Can Annotate Protein Interactions with 70% precision • Tested the effect of source filtering – 90% precision, but recall dropping to 70%
24 Paper V: Web. Prot • Online Implementation, bigger experiment • Can Annotate Protein Interactions with 70% precision • Tested the effect of source filtering – 90% precision, but recall dropping to 70%
 25 Google as a source nih. gov 239 Pub. Med Central Collection of Journals, Books and MEDLINE jbc. org 196 Biological Chemistry, Journal physiology. org 143 American Physiological Society, Collection of Journals endojournals. org 110 Endocrine Society, Collection of Journals asm. org 83 American Society for Microbiology, Collection of Journals ahajournals. org 71 American Heart Association, Collection of Journals nature. com 69 Nature, same as npgjournals. com, Collection of Journals ingentaconnect. com 55 Ingenta Online Publisher, Collection of Journals aacrjournals. org 55 Cancer Research Journal jimmunol. org 51 Immunology, Journal karger. com 48 Karger Medical and Scientific Publisher, Big Collection of Journals pnas. org 44 National Academy of Sciences USA, Proceedings ac. uk 42 MOLECULAR AND CELLULAR BIOLOGY, Journal bloodjournal. org 40 American Society of Hematology, Blood Journal uam. es 39 Information Hyperlinked over Proteins (i. HOP), Network aspetjournals. org 38 Molecular Pharmacology Journal oxfordjournals. org 33 Human Molecular Genetics Journal blackwell-synergy. com 32 Neurochemistry, Journal jcb. org 32 Cell Biology, Journal biochemj. org 30 Biochemical Journal npgjournals. com 30 Collection including European Molecular Biology Organization Journal 1480 4660 facts total from Web. Prot
25 Google as a source nih. gov 239 Pub. Med Central Collection of Journals, Books and MEDLINE jbc. org 196 Biological Chemistry, Journal physiology. org 143 American Physiological Society, Collection of Journals endojournals. org 110 Endocrine Society, Collection of Journals asm. org 83 American Society for Microbiology, Collection of Journals ahajournals. org 71 American Heart Association, Collection of Journals nature. com 69 Nature, same as npgjournals. com, Collection of Journals ingentaconnect. com 55 Ingenta Online Publisher, Collection of Journals aacrjournals. org 55 Cancer Research Journal jimmunol. org 51 Immunology, Journal karger. com 48 Karger Medical and Scientific Publisher, Big Collection of Journals pnas. org 44 National Academy of Sciences USA, Proceedings ac. uk 42 MOLECULAR AND CELLULAR BIOLOGY, Journal bloodjournal. org 40 American Society of Hematology, Blood Journal uam. es 39 Information Hyperlinked over Proteins (i. HOP), Network aspetjournals. org 38 Molecular Pharmacology Journal oxfordjournals. org 33 Human Molecular Genetics Journal blackwell-synergy. com 32 Neurochemistry, Journal jcb. org 32 Cell Biology, Journal biochemj. org 30 Biochemical Journal npgjournals. com 30 Collection including European Molecular Biology Organization Journal 1480 4660 facts total from Web. Prot
 26 Web. Prot
26 Web. Prot
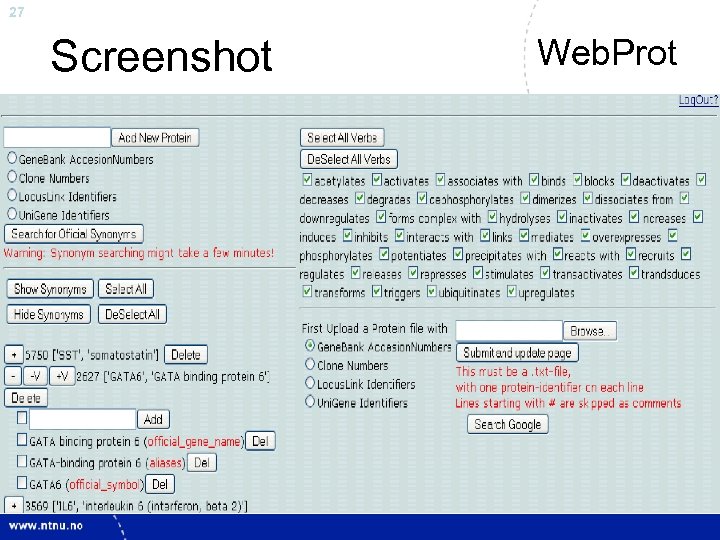 27 Screenshot Web. Prot
27 Screenshot Web. Prot
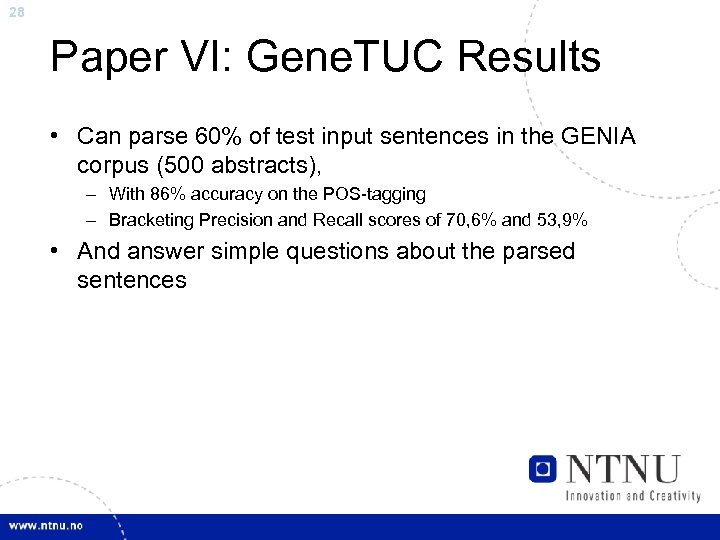 28 Paper VI: Gene. TUC Results • Can parse 60% of test input sentences in the GENIA corpus (500 abstracts), – With 86% accuracy on the POS-tagging – Bracketing Precision and Recall scores of 70, 6% and 53, 9% • And answer simple questions about the parsed sentences
28 Paper VI: Gene. TUC Results • Can parse 60% of test input sentences in the GENIA corpus (500 abstracts), – With 86% accuracy on the POS-tagging – Bracketing Precision and Recall scores of 70, 6% and 53, 9% • And answer simple questions about the parsed sentences
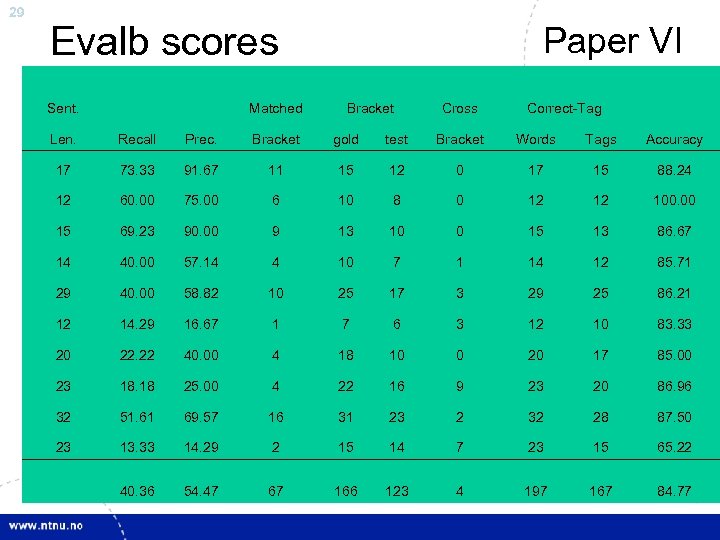 29 Evalb scores Sent. Matched Paper VI Bracket Cross Correct-Tag Len. Recall Prec. Bracket gold test Bracket Words Tags Accuracy 17 73. 33 91. 67 11 15 12 0 17 15 88. 24 12 60. 00 75. 00 6 10 8 0 12 12 100. 00 15 69. 23 90. 00 9 13 10 0 15 13 86. 67 14 40. 00 57. 14 4 10 7 1 14 12 85. 71 29 40. 00 58. 82 10 25 17 3 29 25 86. 21 12 14. 29 16. 67 1 7 6 3 12 10 83. 33 20 22. 22 40. 00 4 18 10 0 20 17 85. 00 23 18. 18 25. 00 4 22 16 9 23 20 86. 96 32 51. 61 69. 57 16 31 23 2 32 28 87. 50 23 13. 33 14. 29 2 15 14 7 23 15 65. 22 40. 36 54. 47 67 166 123 4 197 167 84. 77
29 Evalb scores Sent. Matched Paper VI Bracket Cross Correct-Tag Len. Recall Prec. Bracket gold test Bracket Words Tags Accuracy 17 73. 33 91. 67 11 15 12 0 17 15 88. 24 12 60. 00 75. 00 6 10 8 0 12 12 100. 00 15 69. 23 90. 00 9 13 10 0 15 13 86. 67 14 40. 00 57. 14 4 10 7 1 14 12 85. 71 29 40. 00 58. 82 10 25 17 3 29 25 86. 21 12 14. 29 16. 67 1 7 6 3 12 10 83. 33 20 22. 22 40. 00 4 18 10 0 20 17 85. 00 23 18. 18 25. 00 4 22 16 9 23 20 86. 96 32 51. 61 69. 57 16 31 23 2 32 28 87. 50 23 13. 33 14. 29 2 15 14 7 23 15 65. 22 40. 36 54. 47 67 166 123 4 197 167 84. 77
 30 Summary • 6 papers describing the steps needed to show that Gene. TUC can handle medical text • 60% parsing success-rate may not be enough for a commercial application, – But the fact that it improved from just 10% in 2001 is very promising • Once the parsing success-rate is good enough, Gene. TUC can be tested on Question-Answering – There is a need for a good public dataset that allows measuring and comparing between different QA systems (Future Work)
30 Summary • 6 papers describing the steps needed to show that Gene. TUC can handle medical text • 60% parsing success-rate may not be enough for a commercial application, – But the fact that it improved from just 10% in 2001 is very promising • Once the parsing success-rate is good enough, Gene. TUC can be tested on Question-Answering – There is a need for a good public dataset that allows measuring and comparing between different QA systems (Future Work)
 31 Acknowledgements • Biologists: – Astrid Lægreid, Kamilla Stunes, Kristine Misund, Liv Thommesen, Tonje Strømmen Steigedal • Computer Scientists: – Tore Amble, Arne Halaas, Amund Tveit, Martin Ranang, Harald Søvik, Yoshimasa Tsuruoka, Anders Andenæs, Tor-Kristian Jenssen, Franz Günthner, Jun’ichi Tsujii, Jörg Cassens, Waclaw Kusnierczyk, Tore Bruland, Peep Küngas, Magnus Lie Hetland, Morten Hartman, Hallgeir Bergum, Jo Kristian Bergum, Frode Jünge, Heri Ramampiaro, Rolv Inge Seehuus, Per Kristian Lehre, Clemens Marschner, Petra Maier, Holger Bosk, Sebastian Nagel, Mariya Vitusevych, Yoshimasa Tsuruoka, Jin-Dong Kim, Hong-Woo Chun, Takashi Ninomiya, Yusuke Miyao, Frode Høyvik, Henrik Tveit, Jian Su and others
31 Acknowledgements • Biologists: – Astrid Lægreid, Kamilla Stunes, Kristine Misund, Liv Thommesen, Tonje Strømmen Steigedal • Computer Scientists: – Tore Amble, Arne Halaas, Amund Tveit, Martin Ranang, Harald Søvik, Yoshimasa Tsuruoka, Anders Andenæs, Tor-Kristian Jenssen, Franz Günthner, Jun’ichi Tsujii, Jörg Cassens, Waclaw Kusnierczyk, Tore Bruland, Peep Küngas, Magnus Lie Hetland, Morten Hartman, Hallgeir Bergum, Jo Kristian Bergum, Frode Jünge, Heri Ramampiaro, Rolv Inge Seehuus, Per Kristian Lehre, Clemens Marschner, Petra Maier, Holger Bosk, Sebastian Nagel, Mariya Vitusevych, Yoshimasa Tsuruoka, Jin-Dong Kim, Hong-Woo Chun, Takashi Ninomiya, Yusuke Miyao, Frode Høyvik, Henrik Tveit, Jian Su and others
 32 Questions and Comments • Associate professor Jong C. Park – Computer Science Division, – Korea Advanced Institute of Science and Technology (KAIST), – Daejeon, South Korea • Professor Eivind Hovig – Department of Tumor Biology, – Institute for Cancer Research, – The Norwegian Radium Hospital
32 Questions and Comments • Associate professor Jong C. Park – Computer Science Division, – Korea Advanced Institute of Science and Technology (KAIST), – Daejeon, South Korea • Professor Eivind Hovig – Department of Tumor Biology, – Institute for Cancer Research, – The Norwegian Radium Hospital
 33 Thesis Work • Gene. TUC Project – Use TUC in the Medical Text Domain • Use Google (Bioogle) to Recognize Unknown Entities – Galpha(i 1)-GDP-Al. F(4)(-), Ca 2+, Gastrin • Use Google (Web. Prot) to do Automatic Annotation – Mapping (Bio. Creative): • From Gene/Protein Set of Gene. Ontology Terms
33 Thesis Work • Gene. TUC Project – Use TUC in the Medical Text Domain • Use Google (Bioogle) to Recognize Unknown Entities – Galpha(i 1)-GDP-Al. F(4)(-), Ca 2+, Gastrin • Use Google (Web. Prot) to do Automatic Annotation – Mapping (Bio. Creative): • From Gene/Protein Set of Gene. Ontology Terms
 34 Motivation • Natural language is natural – Talking computers – Voice as input • Repetitive tasks should be automated! – Information Extraction is trivial, if you know what to look for
34 Motivation • Natural language is natural – Talking computers – Voice as input • Repetitive tasks should be automated! – Information Extraction is trivial, if you know what to look for
 35 0: Gene. TUC Diploma Work • NLU Review 2002 – GENIA: HPSG – Park et al. : CCG-parsing • Numbers?
35 0: Gene. TUC Diploma Work • NLU Review 2002 – GENIA: HPSG – Park et al. : CCG-parsing • Numbers?
 36 Paper I: Local Grammars • Maurice Gross: – there is more than 10^50 ways to build a sentence with at most twenty words* * Gross (1997). Construction of Local Grammars
36 Paper I: Local Grammars • Maurice Gross: – there is more than 10^50 ways to build a sentence with at most twenty words* * Gross (1997). Construction of Local Grammars
 37 Paper II: Prot. Chew • Protein Names – Galpha(i 1)-GDP-Al. F(4)(-) – Gastrin – … • Idea: Automatic Extraction – Based on existing dictionaries and machine learning • Results?
37 Paper II: Prot. Chew • Protein Names – Galpha(i 1)-GDP-Al. F(4)(-) – Gastrin – … • Idea: Automatic Extraction – Based on existing dictionaries and machine learning • Results?
![38 evalb • [4] OUTPUT FORMAT FROM THE SCORER • • The scorer gives 38 evalb • [4] OUTPUT FORMAT FROM THE SCORER • • The scorer gives](https://present5.com/presentation/bac4863f446c8e79229af4cd0ab9960a/image-38.jpg) 38 evalb • [4] OUTPUT FORMAT FROM THE SCORER • • The scorer gives individual scores for each sentence, for example: • Sent. Matched Bracket Cross Correct Tag ID Len. Stat. Recal Prec. Bracket gold test Bracket Words Tags Accracy ====================== 1 8 0 100. 00 5 5 5 0 6 5 83. 33 • • • At the end of the output the === Summary === section gives statistics for all sentences, and for sentences <=40 words in length. The summary contains the following information: i) Number of sentences -- total number of sentences. ii) Number of Error/Skip sentences -- should both be 0 if there is no problem with the parsed/gold files. iii) Number of valid sentences = Number of sentences - Number of Error/Skip sentences • • • iv) Bracketing recall = (number of correct constituents) -------------------- (number of constituents in the goldfile) • • • v) Bracketing precision = (number of correct constituents) -------------------- (number of constituents in the parsed file) • vi) Complete match = percentaage of sentences where recall and precision are both 100%. vii) Average cross=(#const crossing a goldfile constituen -------------------- (number of sentences) viii) No crossing = percentage of sentences which have 0 crossing brackets. ix) 2 or less crossing = percentage of sentences which have <=2 crossing brackets. x) Tagging accuracy = percentage of correct POS tags (but see [5]. 3 for exact details of what is counted). • •
38 evalb • [4] OUTPUT FORMAT FROM THE SCORER • • The scorer gives individual scores for each sentence, for example: • Sent. Matched Bracket Cross Correct Tag ID Len. Stat. Recal Prec. Bracket gold test Bracket Words Tags Accracy ====================== 1 8 0 100. 00 5 5 5 0 6 5 83. 33 • • • At the end of the output the === Summary === section gives statistics for all sentences, and for sentences <=40 words in length. The summary contains the following information: i) Number of sentences -- total number of sentences. ii) Number of Error/Skip sentences -- should both be 0 if there is no problem with the parsed/gold files. iii) Number of valid sentences = Number of sentences - Number of Error/Skip sentences • • • iv) Bracketing recall = (number of correct constituents) -------------------- (number of constituents in the goldfile) • • • v) Bracketing precision = (number of correct constituents) -------------------- (number of constituents in the parsed file) • vi) Complete match = percentaage of sentences where recall and precision are both 100%. vii) Average cross=(#const crossing a goldfile constituen -------------------- (number of sentences) viii) No crossing = percentage of sentences which have 0 crossing brackets. ix) 2 or less crossing = percentage of sentences which have <=2 crossing brackets. x) Tagging accuracy = percentage of correct POS tags (but see [5]. 3 for exact details of what is counted). • •
 39 Remember • Present one paper at the time • Summary results and related work also in the end Ta med tabeller for parsing, sammenligning med andre etc. Et eksempel på en kompleks setning med gtb treet. Ref tabell. Sammenlign brackets. Ta med webprot screenshot Related work!! Phd pres. Related work. Lexiquest, 40 verbs, hva er fscore? Fra tore: Hvorfor bare 50%. Er det semantikk eller gramatikk som gjør at 50% feiler
39 Remember • Present one paper at the time • Summary results and related work also in the end Ta med tabeller for parsing, sammenligning med andre etc. Et eksempel på en kompleks setning med gtb treet. Ref tabell. Sammenlign brackets. Ta med webprot screenshot Related work!! Phd pres. Related work. Lexiquest, 40 verbs, hva er fscore? Fra tore: Hvorfor bare 50%. Er det semantikk eller gramatikk som gjør at 50% feiler
 40 Dr. Carl-Fredrik Sørensen (50 min, jeg: tid /2) 5 min intro, state-of-the-art 5 min definitions NLU 10 min thesis/papers overview and Research Questions 15 min three themes and contributions. Evaluation of the work 10 min future work Proof of concept. It can be implemented. Next step? Industry. . . Results are trusted Academic. . . Results are validated through understanding the research process. Dennings, proof of concept Research question. . . soon. . Moores law Proof of performance Shift the work to biologists Medline growth graph. Figure. . . Everything is published. Background: http: //www. coli. uni-saarland. de/~hansu/what_is_cl. html Schopenhauer: imagine how clever a vice man would be, if he knew everything in his books. Inter-annotator agreement in gprot, maybe 80 percent precision is enough? !
40 Dr. Carl-Fredrik Sørensen (50 min, jeg: tid /2) 5 min intro, state-of-the-art 5 min definitions NLU 10 min thesis/papers overview and Research Questions 15 min three themes and contributions. Evaluation of the work 10 min future work Proof of concept. It can be implemented. Next step? Industry. . . Results are trusted Academic. . . Results are validated through understanding the research process. Dennings, proof of concept Research question. . . soon. . Moores law Proof of performance Shift the work to biologists Medline growth graph. Figure. . . Everything is published. Background: http: //www. coli. uni-saarland. de/~hansu/what_is_cl. html Schopenhauer: imagine how clever a vice man would be, if he knew everything in his books. Inter-annotator agreement in gprot, maybe 80 percent precision is enough? !


