1 7 5 5 MSU & Skol. Tech



































- Размер: 24.9 Mегабайта
- Количество слайдов: 48
Описание презентации 1 7 5 5 MSU & Skol. Tech по слайдам
 1 7 5 5 MSU & Skol. Tech Replication in bacteria
1 7 5 5 MSU & Skol. Tech Replication in bacteria
 1 7 5 5 Replication Chemistry of replication O N N N HO O-O P N NN N HO. . . O O N H 2 H NO N H 2 N N O NO O OO PO O -O PO O. . . — O O. . . OA T G CO O -O P — O HO template 3 ‘-end deoxynucleoside ‘-triphosphate 5 N
1 7 5 5 Replication Chemistry of replication O N N N HO O-O P N NN N HO. . . O O N H 2 H NO N H 2 N N O NO O OO PO O -O PO O. . . — O O. . . OA T G CO O -O P — O HO template 3 ‘-end deoxynucleoside ‘-triphosphate 5 N
 1 7 5 5 Replication Chemistry of replication. O N N NH O O — O P N N O. . . O O NH 2 HN O N H 2 N N O O O O PO O — O PO O. . . -O O. . . O A T G C O O- OP O O — OP — O HO template 3 ‘-end- pyrophosphate N
1 7 5 5 Replication Chemistry of replication. O N N NH O O — O P N N O. . . O O NH 2 HN O N H 2 N N O O O O PO O — O PO O. . . -O O. . . O A T G C O O- OP O O — OP — O HO template 3 ‘-end- pyrophosphate N
 1 7 5 5 Replication Chemistry of replication 5′ 3′ OH ppp 5′ 3′ OH 1. Only ‘-end hydroxyl of DNA primer is extended Deoxynucleoside ‘-triphosphate is used Complementary d. NTP is used 3 ( ) 2. -5 3. 4. Pyrophosphate is a leaving group pp = 3’ OH
1 7 5 5 Replication Chemistry of replication 5′ 3′ OH ppp 5′ 3′ OH 1. Only ‘-end hydroxyl of DNA primer is extended Deoxynucleoside ‘-triphosphate is used Complementary d. NTP is used 3 ( ) 2. -5 3. 4. Pyrophosphate is a leaving group pp = 3’ OH
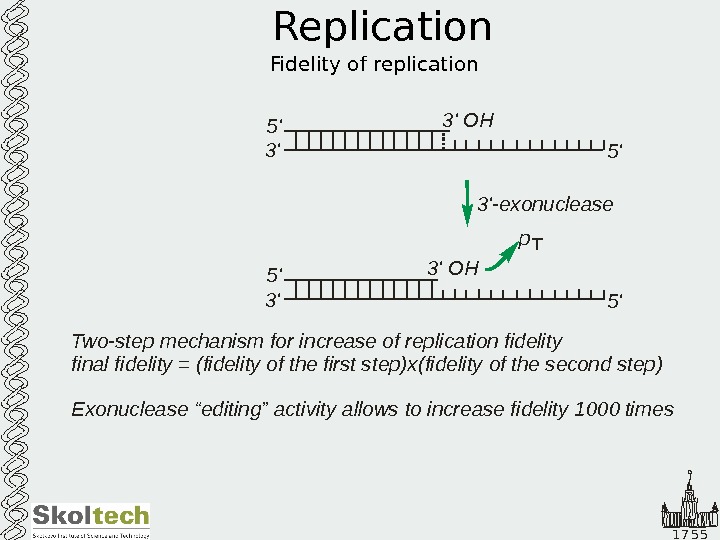
 1 7 5 5 Replication Fidelity of replication 5 ‘ 3’ 5 ‘3’ OH p 3′ exonuclease -5 ‘ 3’ 5 ‘3’ OH Two-step mechanism for increase of replication fidelity final fidelity (fidelity of the first step)x Exonuclease “editing” activity allows to increase fidelity times = 1000 (fidelity of the second step)
1 7 5 5 Replication Fidelity of replication 5 ‘ 3’ 5 ‘3’ OH p 3′ exonuclease -5 ‘ 3’ 5 ‘3’ OH Two-step mechanism for increase of replication fidelity final fidelity (fidelity of the first step)x Exonuclease “editing” activity allows to increase fidelity times = 1000 (fidelity of the second step)
 1 7 5 5 Replication Enzymology of replicationfingers thumb palm exonuclease primer NTP Asp M 2+ magnesium ions coordinate ‘-OH and triphosphate
1 7 5 5 Replication Enzymology of replicationfingers thumb palm exonuclease primer NTP Asp M 2+ magnesium ions coordinate ‘-OH and triphosphate
 1 7 5 5 Replication Enzymology of replication Replicative DNA polymerases DNA polymerase I + 3′ exo + 5′ exo 103 k. D DNA polymerase III 130 k. D polymerase 27 k. D 3′-exo 9 k. D 39 k. D 17 k. D 15 k. D 48 k. D 41 k. D processivity factor 71 k.
1 7 5 5 Replication Enzymology of replication Replicative DNA polymerases DNA polymerase I + 3′ exo + 5′ exo 103 k. D DNA polymerase III 130 k. D polymerase 27 k. D 3′-exo 9 k. D 39 k. D 17 k. D 15 k. D 48 k. D 41 k. D processivity factor 71 k.
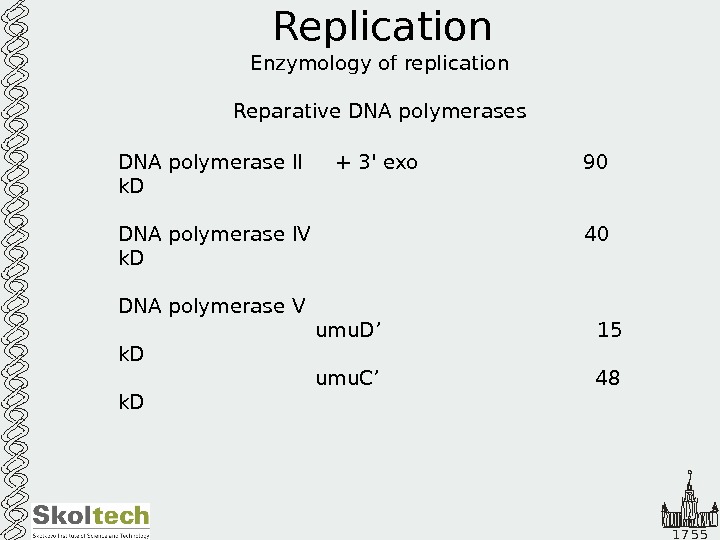
 1 7 5 5 Replication Enzymology of replication Reparative DNA polymerases DNA polymerase II + 3′ exo 90 k. D DNA polymerase IV 40 k. D DNA polymerase V umu. D’ 15 k. D umu. C’ 48 k.
1 7 5 5 Replication Enzymology of replication Reparative DNA polymerases DNA polymerase II + 3′ exo 90 k. D DNA polymerase IV 40 k. D DNA polymerase V umu. D’ 15 k. D umu. C’ 48 k.
 1 7 5 5 Replication fork leading strand lagging strandtemplate Okazaki fragments RNA primers
1 7 5 5 Replication fork leading strand lagging strandtemplate Okazaki fragments RNA primers
 1 7 5 5 Replication helicase SSB primase 5 ‘ exo RNase Н ligase RNase Н ligaseligase. Replication fork
1 7 5 5 Replication helicase SSB primase 5 ‘ exo RNase Н ligase RNase Н ligaseligase. Replication fork
 1 7 5 5 Replication Helicases helicase 5′ — 3′ helicase 3 5 ‘ — ‘ helicase Dna. B 52 k. D main replicative helicase Rep k. D replicative, reparative, needed for replication fork restart 77 Rec. B Rec. C Rec. D Rec. G Rec. Q Uvr. D helicase IV recombination and reparation. ATP ADP
1 7 5 5 Replication Helicases helicase 5′ — 3′ helicase 3 5 ‘ — ‘ helicase Dna. B 52 k. D main replicative helicase Rep k. D replicative, reparative, needed for replication fork restart 77 Rec. B Rec. C Rec. D Rec. G Rec. Q Uvr. D helicase IV recombination and reparation. ATP ADP
 1 7 5 5 Replication Helicases Dna. B 6 Rep 2 ope n close d
1 7 5 5 Replication Helicases Dna. B 6 Rep 2 ope n close d
 1 7 5 5 Replication Helicases Working cycle of Rep 2 Translocation Unwinding
1 7 5 5 Replication Helicases Working cycle of Rep 2 Translocation Unwinding
 1 7 5 5 Replication form components SSB tetramer k. D on DNA may form octamers, covering b, spacer b. , 19 145 30 binds ss. DNA and assists all replication machinerybinds ss. DNA and assists all replication machinery
1 7 5 5 Replication form components SSB tetramer k. D on DNA may form octamers, covering b, spacer b. , 19 145 30 binds ss. DNA and assists all replication machinerybinds ss. DNA and assists all replication machinery
 1 7 5 5 Replication Primase. Dna. G 66 k. D primase makes RNA primers and interact with Dna. B (Dna. G Dna. B ) prefers YYR patches and is displaced by DNA polymerase III 3 6 helicaseprimase Dna. B 6 Dna. G
1 7 5 5 Replication Primase. Dna. G 66 k. D primase makes RNA primers and interact with Dna. B (Dna. G Dna. B ) prefers YYR patches and is displaced by DNA polymerase III 3 6 helicaseprimase Dna. B 6 Dna. G
 1 7 5 5 Replication Primase
1 7 5 5 Replication Primase
 1 7 5 5 Replication Removal of primer 5 ‘-exo activity of DNA polymerase I removes the last nucleotides of primer RNase k. D cleaves RNA in a RNA-DNA duplex Н 18 Removes RNA left after transcription to prevent replication start at random positions
1 7 5 5 Replication Removal of primer 5 ‘-exo activity of DNA polymerase I removes the last nucleotides of primer RNase k. D cleaves RNA in a RNA-DNA duplex Н 18 Removes RNA left after transcription to prevent replication start at random positions
 1 7 5 5 Replication Ligation of Okazaki fragments Lig. A 4 k. D Lig. B 63 k. D E. coli ligase use NAD T 4 phage and eukaryotic ligase use ATP 7 +ATP AMP + PPi p
1 7 5 5 Replication Ligation of Okazaki fragments Lig. A 4 k. D Lig. B 63 k. D E. coli ligase use NAD T 4 phage and eukaryotic ligase use ATP 7 +ATP AMP + PPi p
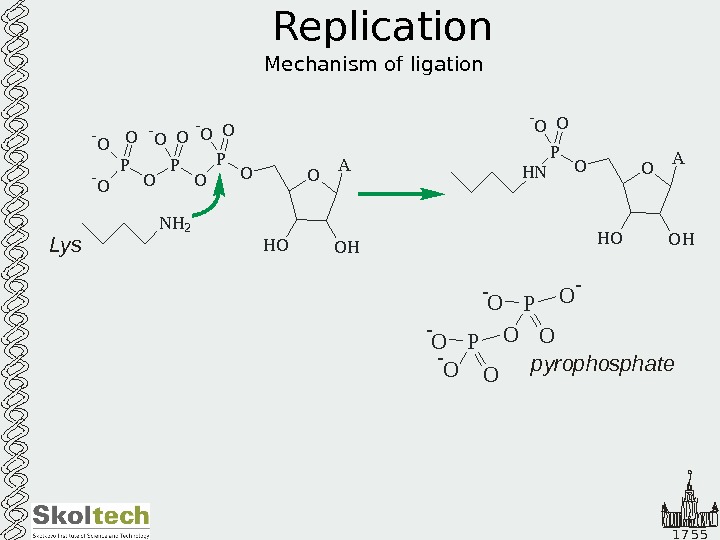
 1 7 5 5 Replication Mechanism of ligation O A O HH OOP O OP- O O — O N H 2 O A O HH OOP H N O- O Lys O O- O P — O — pyrophosphate
1 7 5 5 Replication Mechanism of ligation O A O HH OOP O OP- O O — O N H 2 O A O HH OOP H N O- O Lys O O- O P — O — pyrophosphate
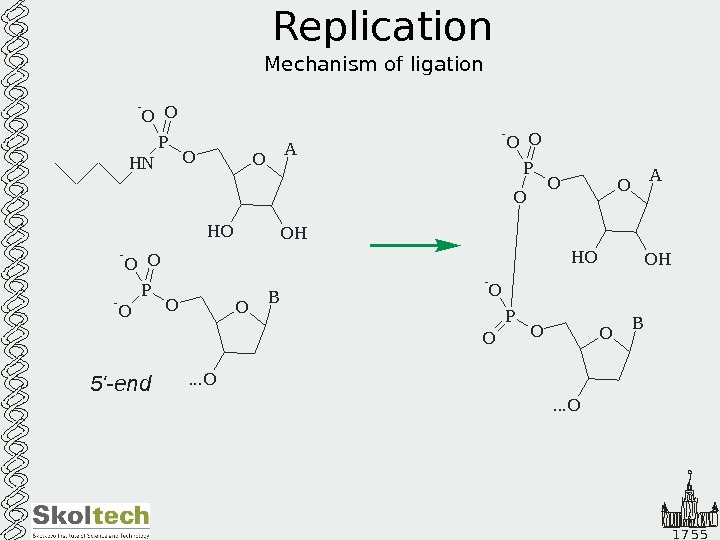
 1 7 5 5 Replication Mechanism of ligation O A O HH OOP H N O- O O B. . . OOP — O O A O HH OOP O- O O O B. . . OOP O — O 5′-end
1 7 5 5 Replication Mechanism of ligation O A O HH OOP H N O- O O B. . . OOP — O O A O HH OOP O- O O O B. . . OOP O — O 5′-end
 1 7 5 5 Replication Mechanism of ligation O A O HH OOP O- O O O B. . . OOP O — OO B H OOP — O O A O HH OOP — O O B H OOP O O- O- O O — O P O B O 5′-end 3′-end AMP ligated DNA fragments
1 7 5 5 Replication Mechanism of ligation O A O HH OOP O- O O O B. . . OOP O — OO B H OOP — O O A O HH OOP — O O B H OOP O O- O- O O — O P O B O 5′-end 3′-end AMP ligated DNA fragments
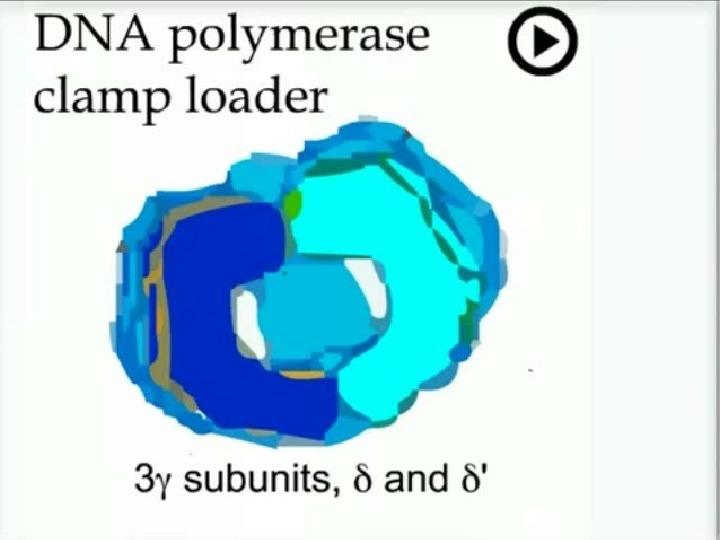
 1 7 5 5 Replication DNA polymerase III Core 130 k. D 27 k. D ‘- 9 k. D 3 k. D 48 k. D 72 polymerase exo Clamp (processivity factor) 2 x 41 k. D Clamp loader 3 k. D 17 k. D 15 k. D 3 9 2 ’ 71 2 x k. D dimerisation X 2} X 2 } coded by a single gene
1 7 5 5 Replication DNA polymerase III Core 130 k. D 27 k. D ‘- 9 k. D 3 k. D 48 k. D 72 polymerase exo Clamp (processivity factor) 2 x 41 k. D Clamp loader 3 k. D 17 k. D 15 k. D 3 9 2 ’ 71 2 x k. D dimerisation X 2} X 2 } coded by a single gene
 1 7 5 5 Replication Processivity factor (clamp) Processivity factor, clamp, has pseudo 6 -fold symmetry. In E. coli -clamp is a dimer of 3 -domain proteins Т 4 phage and eukariotes contain trimer of 2 -domain proteins (PCNA)
1 7 5 5 Replication Processivity factor (clamp) Processivity factor, clamp, has pseudo 6 -fold symmetry. In E. coli -clamp is a dimer of 3 -domain proteins Т 4 phage and eukariotes contain trimer of 2 -domain proteins (PCNA)
 1 7 5 5 Replication DNA polymerase III leading strand lagging strand core 2 2 leading strand lagging strand core
1 7 5 5 Replication DNA polymerase III leading strand lagging strand core 2 2 leading strand lagging strand core
 1 7 5 5 Replication Clamp loading 2 ’+ ATP ATP complex 5 ‘ 5 ‘ ADPADP / ’ core
1 7 5 5 Replication Clamp loading 2 ’+ ATP ATP complex 5 ‘ 5 ‘ ADPADP / ’ core
 1 7 5 5 Replication Clamp loading
1 7 5 5 Replication Clamp loading
 1 7 5 5 Replication Chemistry of replication helicase primase leading starnd lagging strandparental DNA core 2 2 ’
1 7 5 5 Replication Chemistry of replication helicase primase leading starnd lagging strandparental DNA core 2 2 ’
 1 7 5 5 Replication Supercoiling problem ahead of replication fork parental DN
1 7 5 5 Replication Supercoiling problem ahead of replication fork parental DN
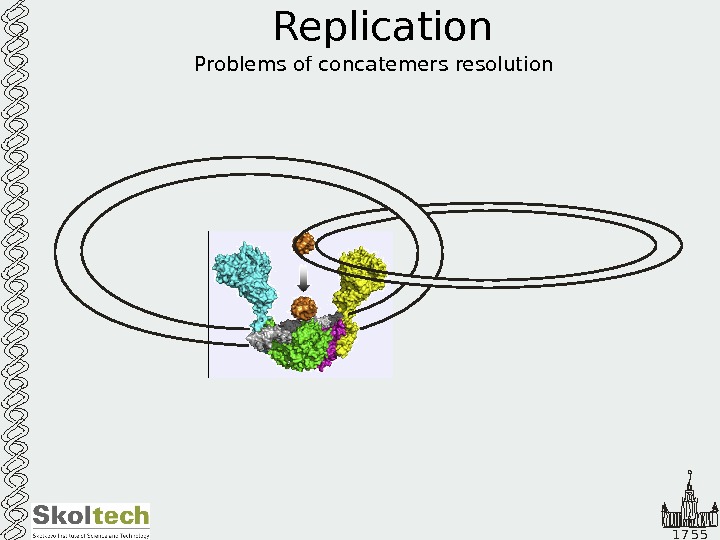
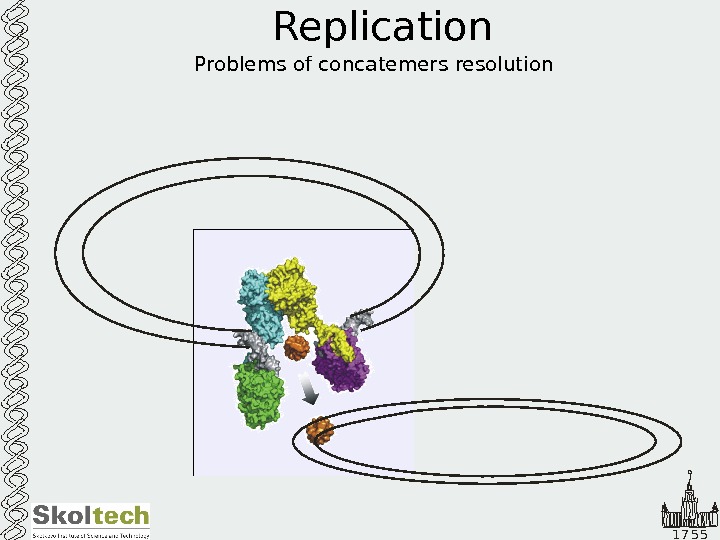
 1 7 5 5 Replication Problems of concatemers resolution
1 7 5 5 Replication Problems of concatemers resolution
 1 7 5 5 Replication Problems of concatemers resolution
1 7 5 5 Replication Problems of concatemers resolution
 1 7 5 5 Replication Problems of concatemers resolution
1 7 5 5 Replication Problems of concatemers resolution
 1 7 5 5 Replication Similar supercoiling problems are relevant for transcription DNA RNA polymerase
1 7 5 5 Replication Similar supercoiling problems are relevant for transcription DNA RNA polymerase
 1 7 5 5 Replication Origin of replication 245 bp DNA unwinding site Dna. A binding sites IHF binding site Fis binding site dam methylation sites GATC dam methylation 5′-GATC CTAG-5′ NN N NH N M e
1 7 5 5 Replication Origin of replication 245 bp DNA unwinding site Dna. A binding sites IHF binding site Fis binding site dam methylation sites GATC dam methylation 5′-GATC CTAG-5′ NN N NH N M e
 1 7 5 5 Replication Initiation of replication is regulated by methylation of dam sites in ori region Dna. A*ADP Seq. A semimethylated dam sites binds Seq. A inhibitor protein initiation of replication can not start before the completion of methylation
1 7 5 5 Replication Initiation of replication is regulated by methylation of dam sites in ori region Dna. A*ADP Seq. A semimethylated dam sites binds Seq. A inhibitor protein initiation of replication can not start before the completion of methylation
 1 7 5 5 Replication Initiation of replication Dna. A*ADP Seq. A Fis IHFFis and IHF bends DNA Binding of Dna. A ATP * Dna. A*ATPDNA unwinding at initiation site
1 7 5 5 Replication Initiation of replication Dna. A*ADP Seq. A Fis IHFFis and IHF bends DNA Binding of Dna. A ATP * Dna. A*ATPDNA unwinding at initiation site
 1 7 5 5 Replication Chemistry of replication. Dna. B Dna. C 6 6 helicase binding 6 x ATP 6 x ADP, Dna. C Dna. B Dna. C 6 6 Dna. G primase RNA primer. SSBDna. B Dna. C 6 6 Dna. G primase RNA primer. SSBSSBSSBSSBSSBSS
1 7 5 5 Replication Chemistry of replication. Dna. B Dna. C 6 6 helicase binding 6 x ATP 6 x ADP, Dna. C Dna. B Dna. C 6 6 Dna. G primase RNA primer. SSBDna. B Dna. C 6 6 Dna. G primase RNA primer. SSBSSBSSBSSBSSBSS
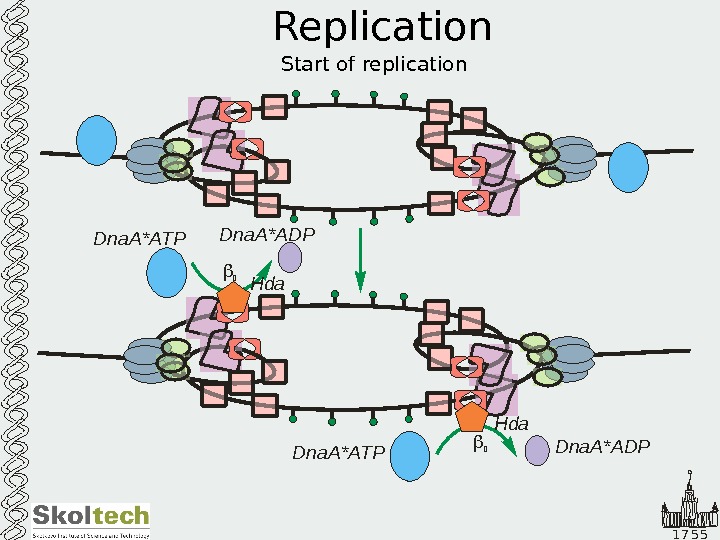
 1 7 5 5 Replication Start of replication Dna. A*ATP Dna. A*ADPDna. A*ADP Dna. A*ATP 2 2 Hda. Hda Dna. A*ATP Dna. A*ADPDna. A*ADP Dna. A*ATP 2 2 Hda. Hda Dna. A*ATP Dna. A*ADPDna. A*ADP Dna. A*ATP 2 2 Hda. Hda Dna. A*ATP Dna. A*ADPDna. A*ADP Dna. A*ATP 2 2 Hda Dna. A*ATP Dna. A*ADP Dna. A*ATP 2 2 Hda
1 7 5 5 Replication Start of replication Dna. A*ATP Dna. A*ADPDna. A*ADP Dna. A*ATP 2 2 Hda. Hda Dna. A*ATP Dna. A*ADPDna. A*ADP Dna. A*ATP 2 2 Hda. Hda Dna. A*ATP Dna. A*ADPDna. A*ADP Dna. A*ATP 2 2 Hda. Hda Dna. A*ATP Dna. A*ADPDna. A*ADP Dna. A*ATP 2 2 Hda Dna. A*ATP Dna. A*ADP Dna. A*ATP 2 2 Hda
 1 7 5 5 Replication Termination of replicationori. C [ dif. Ter. C Ter. B Ter. F Ter. G Ter. J Ter. H Ter. I Ter. E Ter. D Ter. A ori.
1 7 5 5 Replication Termination of replicationori. C [ dif. Ter. C Ter. B Ter. F Ter. G Ter. J Ter. H Ter. I Ter. E Ter. D Ter. A ori.
 1 7 5 5 Replication Termination of replication protein Tus stops Dna. B helicase approaching from one side, but not the other GO!
1 7 5 5 Replication Termination of replication protein Tus stops Dna. B helicase approaching from one side, but not the other GO!
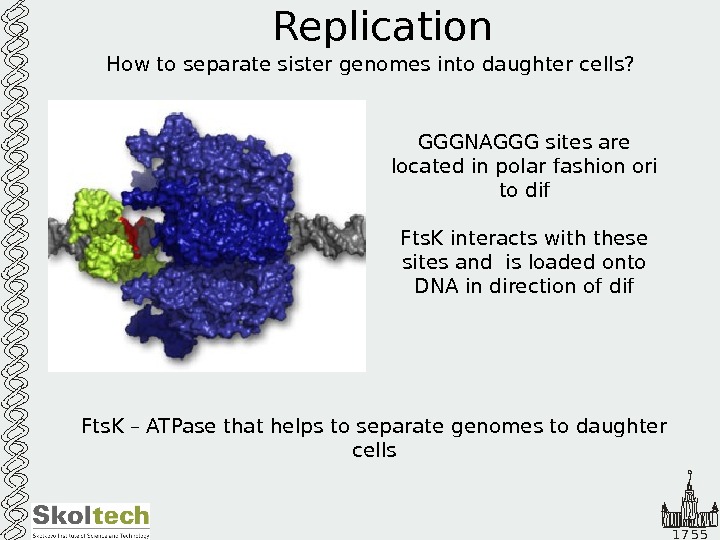
 1 7 5 5 Replication How to separate sister genomes into daughter cells? Fts. K – ATPase that helps to separate genomes to daughter cells GGGNAGGG sites are located in polar fashion ori to dif Fts. K interacts with these sites and is loaded onto DNA in direction of dif
1 7 5 5 Replication How to separate sister genomes into daughter cells? Fts. K – ATPase that helps to separate genomes to daughter cells GGGNAGGG sites are located in polar fashion ori to dif Fts. K interacts with these sites and is loaded onto DNA in direction of dif
 1 7 5 5 Replication Problem of genome dimers Fts. K – ATPase that helps to separate genomes to daughter cells ori. C dif Fts. K ATPADPFts. KATP ADP
1 7 5 5 Replication Problem of genome dimers Fts. K – ATPase that helps to separate genomes to daughter cells ori. C dif Fts. K ATPADPFts. KATP ADP
 1 7 5 5 Replication Problem of genome dimers Xer. C/Xer. D – recombinase is attracted to dif sites by Fts. K ori. C dif Fts. K
1 7 5 5 Replication Problem of genome dimers Xer. C/Xer. D – recombinase is attracted to dif sites by Fts. K ori. C dif Fts. K
 1 7 5 5 Replication Problem of genome dimers ori. C dif Fts. K ATP ADPFts. KATP ADP Xer. C/Xer. D – recombinase ori. C dif Fts. Kori. C difdif
1 7 5 5 Replication Problem of genome dimers ori. C dif Fts. K ATP ADPFts. KATP ADP Xer. C/Xer. D – recombinase ori. C dif Fts. Kori. C difdif
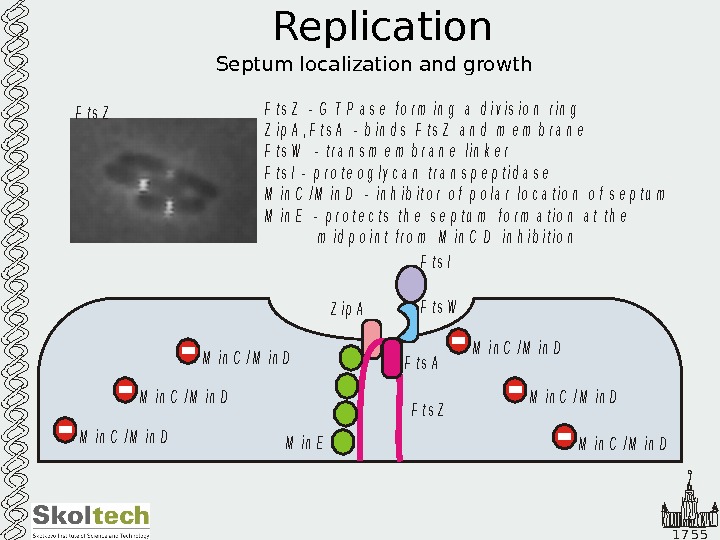
 1 7 5 5 Replication Septum localization and growth. F t s Z Z i p A F t s W F t s I F t s Z G T P a s e f o r m i n g a d i v i s i o n r i n g Z i p A , F t s A — b i n d s F t s Z a n d m e m b r a n e F t s W — t r a n s m e m b r a n e l i n k e r F t s I — p r o t e o g l y c a n t r a n s p e p t i d a s e M i n C / M i n D — i n h i b i t o r o f p o l a r l o c a t i o n o f s e p t u m M i n E — p r o t e c t s t h e s e p t u m f o r m a t i o n a t t h e m i d p o i n t f r o m M i n C D i n h i b i t i o n — M i n C / M i n D M i n C / M i n D M i n
1 7 5 5 Replication Septum localization and growth. F t s Z Z i p A F t s W F t s I F t s Z G T P a s e f o r m i n g a d i v i s i o n r i n g Z i p A , F t s A — b i n d s F t s Z a n d m e m b r a n e F t s W — t r a n s m e m b r a n e l i n k e r F t s I — p r o t e o g l y c a n t r a n s p e p t i d a s e M i n C / M i n D — i n h i b i t o r o f p o l a r l o c a t i o n o f s e p t u m M i n E — p r o t e c t s t h e s e p t u m f o r m a t i o n a t t h e m i d p o i n t f r o m M i n C D i n h i b i t i o n — M i n C / M i n D M i n C / M i n D M i n
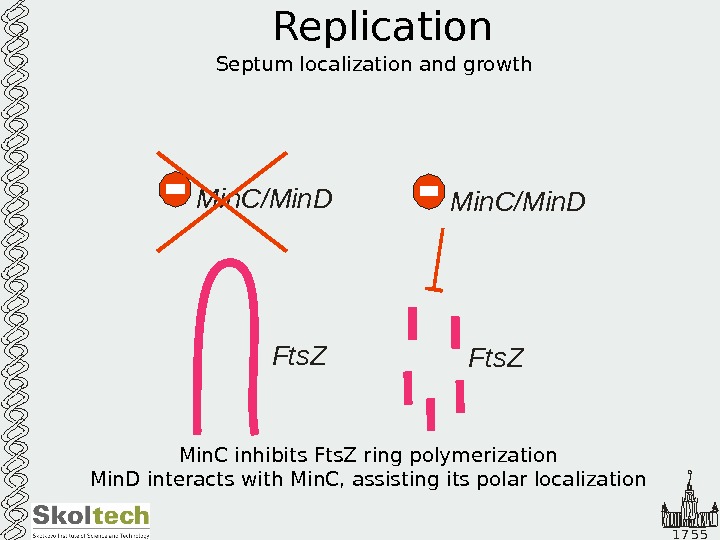
 1 7 5 5 Replication Septum localization and growth Min. C/Min. D Fts. Z Min. C inhibits Fts. Z ring polymerization Min. D interacts with Min. C, assisting its polar localization
1 7 5 5 Replication Septum localization and growth Min. C/Min. D Fts. Z Min. C inhibits Fts. Z ring polymerization Min. D interacts with Min. C, assisting its polar localization
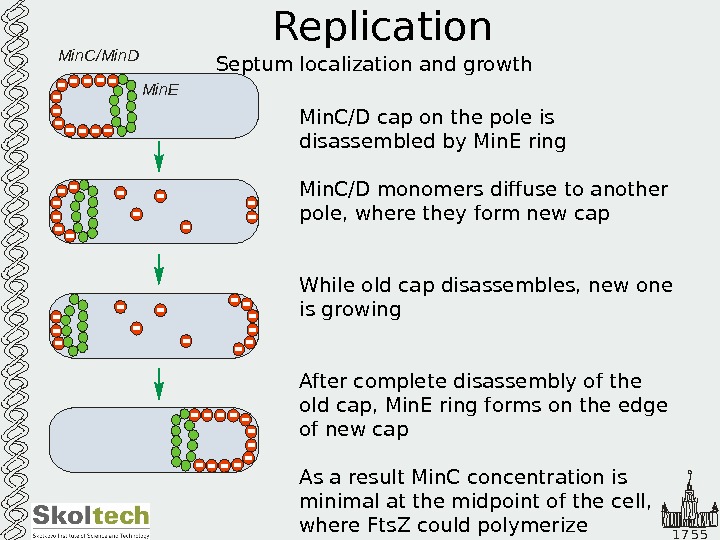
 1 7 5 5 Replication Septum localization and growth Min. C/D cap on the pole is disassembled by Min. E ring Min. C/D monomers diffuse to another pole, where they form new cap While old cap disassembles, new one is growing After complete disassembly of the old cap, Min. E ring forms on the edge of new cap As a result Min. C concentration is minimal at the midpoint of the cell, where Fts. Z could polymerize. Min. C/Min. D Min.
1 7 5 5 Replication Septum localization and growth Min. C/D cap on the pole is disassembled by Min. E ring Min. C/D monomers diffuse to another pole, where they form new cap While old cap disassembles, new one is growing After complete disassembly of the old cap, Min. E ring forms on the edge of new cap As a result Min. C concentration is minimal at the midpoint of the cell, where Fts. Z could polymerize. Min. C/Min. D Min.
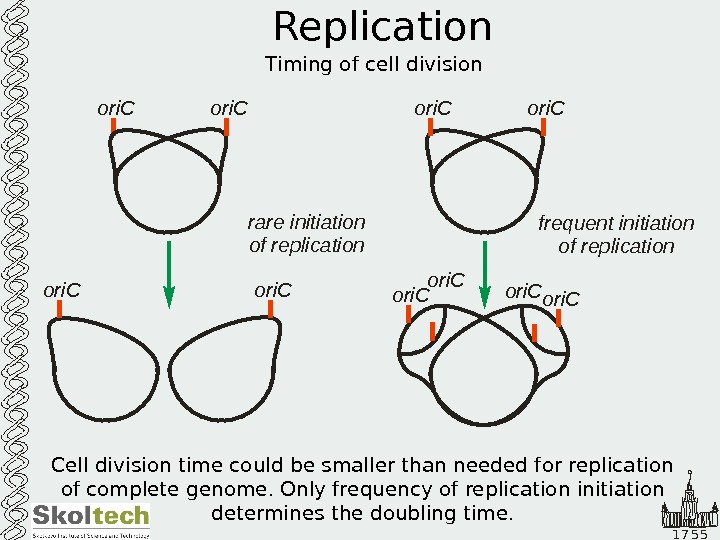
 1 7 5 5 Replication Timing of cell divisionori. C rare initiation of replication frequent initiation of replication ori. Cori. C ori. C Cell division time could be smaller than needed for replication of complete genome. Only frequency of replication initiation determines the doubling time.
1 7 5 5 Replication Timing of cell divisionori. C rare initiation of replication frequent initiation of replication ori. Cori. C ori. C Cell division time could be smaller than needed for replication of complete genome. Only frequency of replication initiation determines the doubling time.
 1 7 5 5 Replication Several factors contribute to regulation of initiation of replication 1. Time to methylate dam sites of ori. C Dna. A Seq. A 2 subunit of DNAP III and Hda ATP hydrolysis on Dna. A. 2 inhibit initiation of replication by stimulation of Dna. A*ATPDna. A*ADP 2 Hda 3. Promoter region of Dna. A contains dam sites and is inhibited by Seq. A Pdna.
1 7 5 5 Replication Several factors contribute to regulation of initiation of replication 1. Time to methylate dam sites of ori. C Dna. A Seq. A 2 subunit of DNAP III and Hda ATP hydrolysis on Dna. A. 2 inhibit initiation of replication by stimulation of Dna. A*ATPDna. A*ADP 2 Hda 3. Promoter region of Dna. A contains dam sites and is inhibited by Seq. A Pdna.

