06328f69ba4728f48eb8dcb077464bc9.ppt
- Количество слайдов: 1
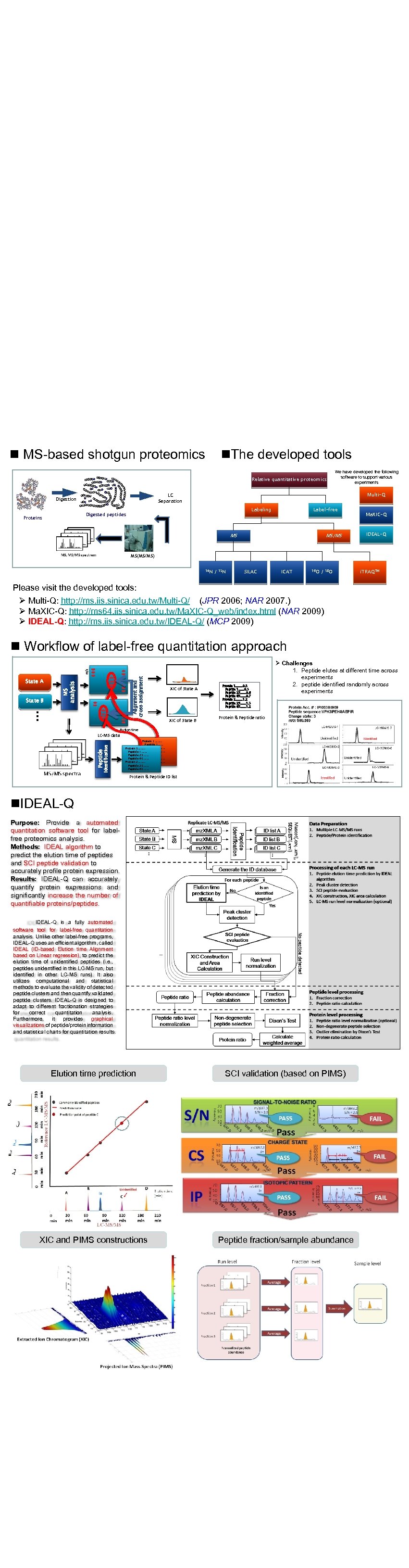
自動化蛋白質定量系統 Automatic Protein Quantitation System n MS-based shotgun proteomics A A D M N A Digestion L T Y V E E P E G A F L G D H H E F A D L E M M G H T P H R I E D P V P G K F H K K L L S A D N Q S H A T Q L L K G V M I S I A K A T E L S V T N G Y H Multi-Q LC Separation A Q L L E W Q G G V L A I I I E L F Labeling Digested peptides Proteins We have developed the following software to support various experiments D S L G W R G A K K K L G K V E GV Relative quantitative proteomics D F H K M E G G G Q E K L F K K A L K Q G M K E S E L D V K F E S A H G L K n. The developed tools Label-free MS MS/MS Ma. XIC-Q IDEAL-Q MS(MS/MS) 14 N / 15 N SILAC ICAT 16 O / 18 O i. TRAQTM Please visit the developed tools: Ø Multi-Q: http: //ms. iis. sinica. edu. tw/Multi-Q/ (JPR 2006; NAR 2007. ) Ø Ma. XIC-Q: http: //ms 64. iis. sinica. edu. tw/Ma. XIC-Q_web/index. html (NAR 2009) Ø IDEAL-Q: http: //ms. iis. sinica. edu. tw/IDEAL-Q/ (MCP 2009) MS analysis State A Alignment and cross assignment m/z n Workflow of label-free quantitation approach State B XIC of State A … XIC of State B Protein 1……. . 0. 5 Peptide 1…… 0. 3 Peptide 25…… 0. 6 Peptide 65…… 0. 7 Protein 2……. . 1. 2 Peptide 25…… 1. 3 Peptide 65…… 1. 2 Ø Challenges 1. Peptide elutes at different time across experiments 2. peptide identified randomly across experiments Protein & Peptide ratio Elution time Peptide identification LC-MS data Protein 1……. . Peptide 1…… Peptide 25…… Protein 5……. . Peptide 65…… Peptide 1…… Protein 2……. . Peptide 25…… Peptide 65…… Protein & Peptide ID list n. IDEAL-Q Purpose: Provide a automated quantitation software tool for labelfree proteomics analysis. Methods: IDEAL algorithm to predict the elution time of peptides and SCI peptide validation to accurately profile protein expression. Results: IDEAL-Q can accurately quantify protein expressions and significantly increase the number of quantifiable proteins/peptides. IDEAL-Q is a fully automated software tool for label-free quantitation analysis. Unlike other label-free programs, IDEAL-Q uses an efficient algorithm, called IDEAL (ID-based Elution time Alignment based on Linear regression), to predict the elution time of unidentified peptides (i. e. , peptides unidentified in this LC-MS run, but identified in other LC-MS runs). It also utilizes computational and statistical methods to evaluate the validity of detected peptide clusters and then quantify validated peptide clusters. IDEAL-Q is designed to adapt to different fractionation strategies for correct quantitation analysis. Furthermore, it provides graphical visualizations of peptide/protein information and statistical charts for quantitation results. Elution time prediction SCI validation (based on PIMS) XIC and PIMS constructions Peptide fraction/sample abundance 生物資訊實驗室 計畫主持人 許聞廉 特聘研究員 宋定懿 研究員
06328f69ba4728f48eb8dcb077464bc9.ppt