Studying Pathogenesis of Hodgkin Lymphomas Sibrand Poppema Department











the_pathology_of_hd.ppt
- Размер: 18.9 Mегабайта
- Количество слайдов: 68
Описание презентации Studying Pathogenesis of Hodgkin Lymphomas Sibrand Poppema Department по слайдам
 Studying Pathogenesis of Hodgkin Lymphomas Sibrand Poppema Department of Pathology & Laboratory Medicine University Medical Center Groningen
Studying Pathogenesis of Hodgkin Lymphomas Sibrand Poppema Department of Pathology & Laboratory Medicine University Medical Center Groningen
 University Medical Center Groningen
University Medical Center Groningen
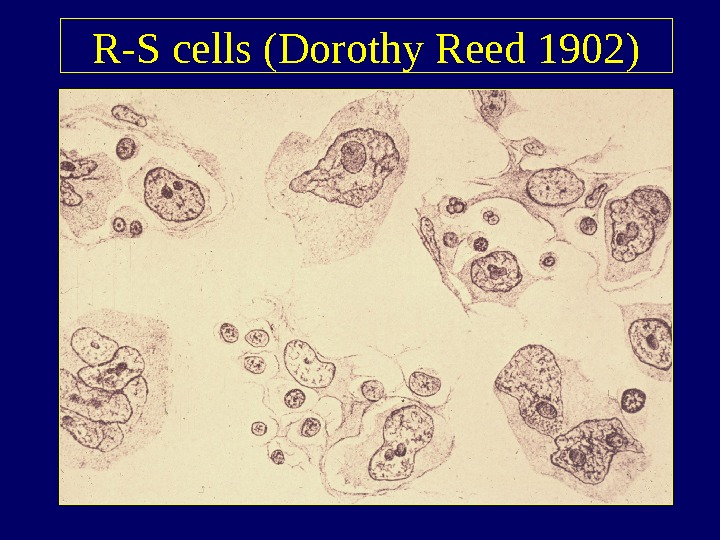
 R-S cells (Dorothy Reed 1902)
R-S cells (Dorothy Reed 1902)
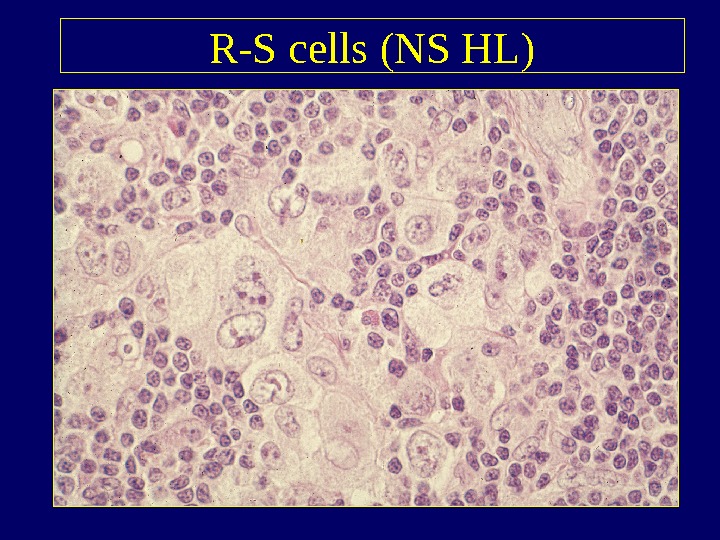
 R-S cells (NS HL)
R-S cells (NS HL)

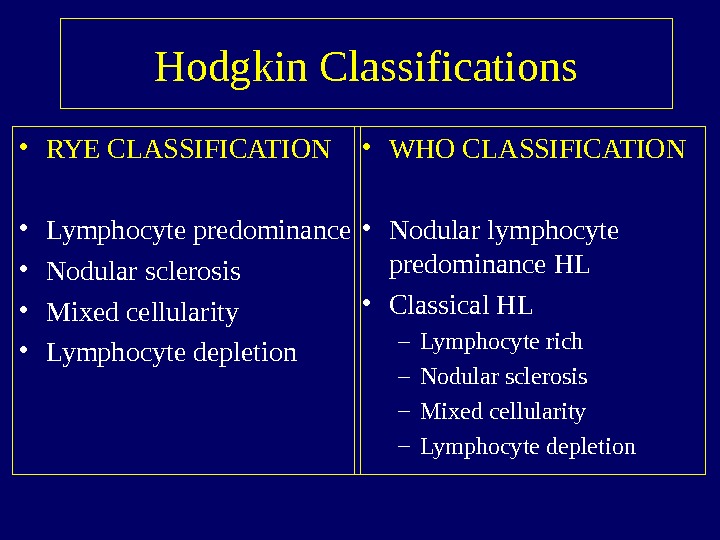
 Hodgkin Classifications • RYE CLASSIFICATION • Lymphocyte predominance • Nodular sclerosis • Mixed cellularity • Lymphocyte depletion • WHO CLASSIFICATION • Nodular lymphocyte predominance HL • Classical HL – Lymphocyte rich – Nodular sclerosis – Mixed cellularity – Lymphocyte depletion
Hodgkin Classifications • RYE CLASSIFICATION • Lymphocyte predominance • Nodular sclerosis • Mixed cellularity • Lymphocyte depletion • WHO CLASSIFICATION • Nodular lymphocyte predominance HL • Classical HL – Lymphocyte rich – Nodular sclerosis – Mixed cellularity – Lymphocyte depletion
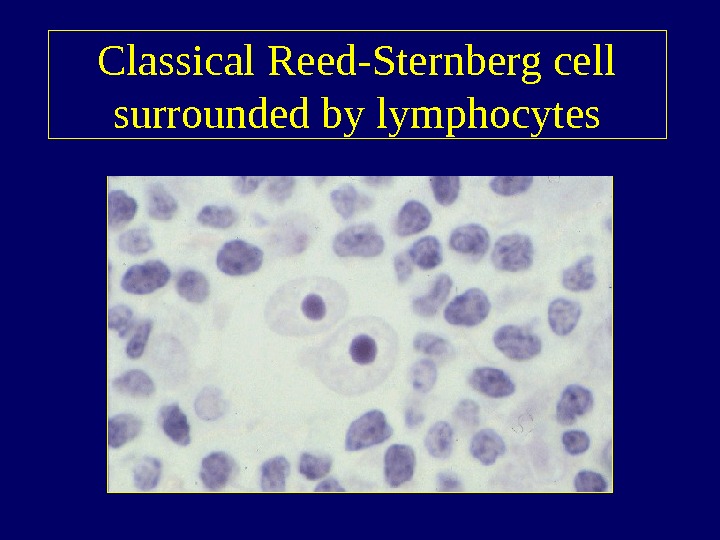
 Classical Reed-Sternberg cell surrounded by lymphocytes
Classical Reed-Sternberg cell surrounded by lymphocytes
 R-S cells and Lymphocytes • Lineage • Clonality • Role of EB virus • Subtype lymphocytes • Cytokines • Chemokines
R-S cells and Lymphocytes • Lineage • Clonality • Role of EB virus • Subtype lymphocytes • Cytokines • Chemokines
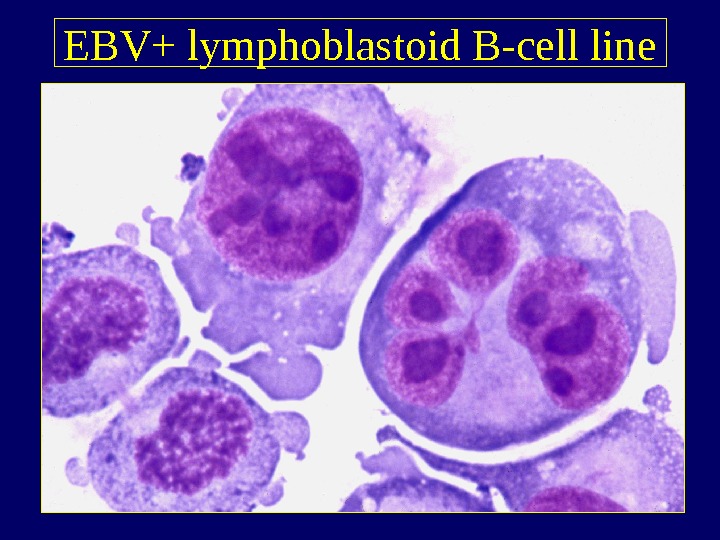
 EBV+ lymphoblastoid B-cell line
EBV+ lymphoblastoid B-cell line
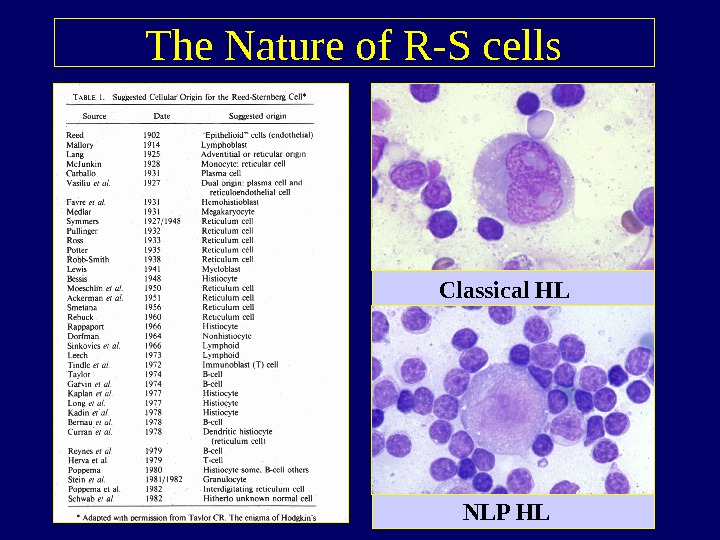
 The Nature of R-S cells Classical HL NLP HL
The Nature of R-S cells Classical HL NLP HL
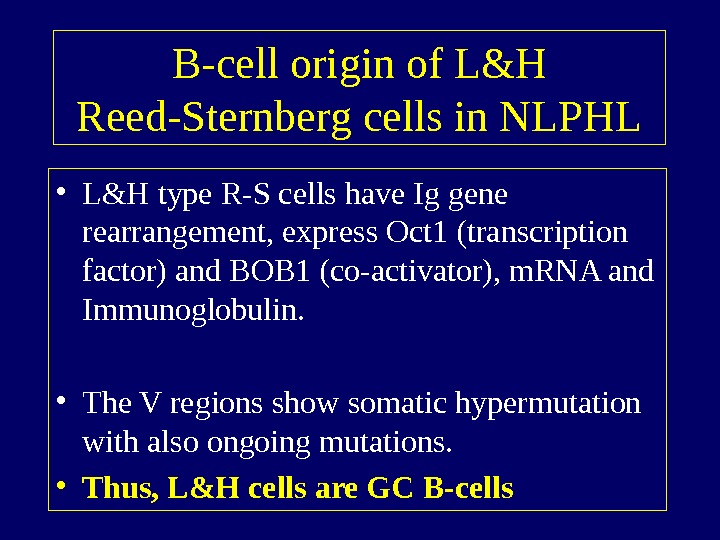
 B-cell origin of L&H Reed-Sternberg cells in NLPHL • L&H type R-S cells have Ig gene rearrangement, express Oct 1 (transcription factor) and BOB 1 (co-activator), m. RNA and Immunoglobulin. • The V regions show somatic hypermutation with also ongoing mutations. • Thus, L&H cells are GC B-cells
B-cell origin of L&H Reed-Sternberg cells in NLPHL • L&H type R-S cells have Ig gene rearrangement, express Oct 1 (transcription factor) and BOB 1 (co-activator), m. RNA and Immunoglobulin. • The V regions show somatic hypermutation with also ongoing mutations. • Thus, L&H cells are GC B-cells
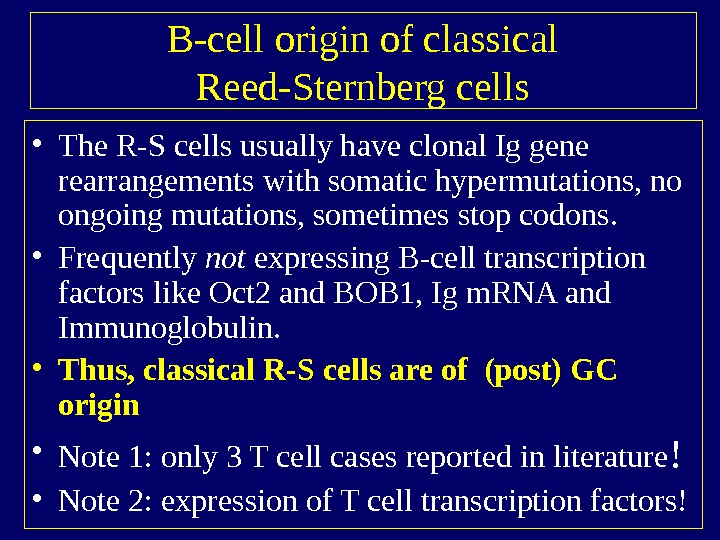
 B-cell origin of classical Reed-Sternberg cells • The R-S cells usually have clonal Ig gene rearrangements with somatic hypermutations, no ongoing mutations, sometimes stop codons. • Frequently not expressing B-cell transcription factors like Oct 2 and BOB 1, Ig m. RNA and Immunoglobulin. • Thus, classical R-S cells are of (post) GC origin • Note 1: only 3 T cell cases reported in literature ! • Note 2: expression of T cell transcription factors!
B-cell origin of classical Reed-Sternberg cells • The R-S cells usually have clonal Ig gene rearrangements with somatic hypermutations, no ongoing mutations, sometimes stop codons. • Frequently not expressing B-cell transcription factors like Oct 2 and BOB 1, Ig m. RNA and Immunoglobulin. • Thus, classical R-S cells are of (post) GC origin • Note 1: only 3 T cell cases reported in literature ! • Note 2: expression of T cell transcription factors!
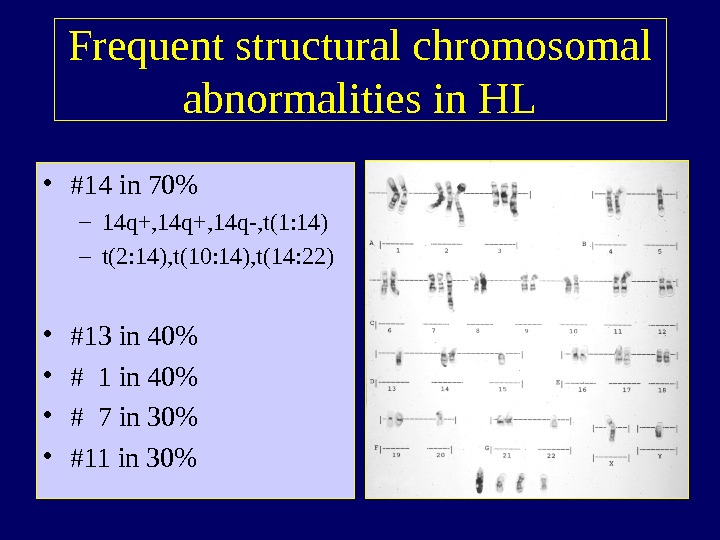
 Frequent structural chromosomal abnormalities in HL • #14 in 70% – 14 q+, 14 q-, t(1: 14) – t(2: 14), t(10: 14), t(14: 22) • #13 in 40% • # 1 in 40% • # 7 in 30% • #11 in 30%
Frequent structural chromosomal abnormalities in HL • #14 in 70% – 14 q+, 14 q-, t(1: 14) – t(2: 14), t(10: 14), t(14: 22) • #13 in 40% • # 1 in 40% • # 7 in 30% • #11 in 30%
 Some questions on HL • Why is there an immune response? • Do R-S cells produce cytokines/chemokines? • Is an antigen involved? • Why is the immune response not effective? • Is the T-cell activation inappropriate? • Is there a problem with antigen presentation? • Is there a bias in T-cell attraction? • Do R-S cells produce immunosuppressive factors?
Some questions on HL • Why is there an immune response? • Do R-S cells produce cytokines/chemokines? • Is an antigen involved? • Why is the immune response not effective? • Is the T-cell activation inappropriate? • Is there a problem with antigen presentation? • Is there a bias in T-cell attraction? • Do R-S cells produce immunosuppressive factors?
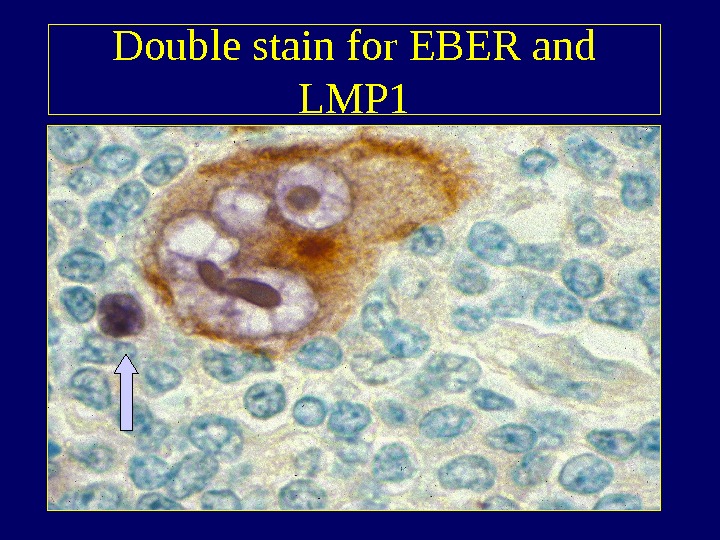
 Double stain for EBER and LMP
Double stain for EBER and LMP
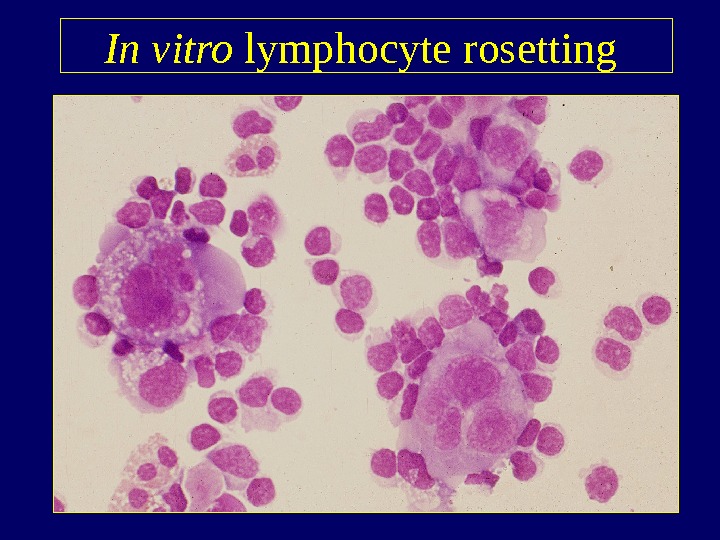
 In vitro lymphocyte rosetting
In vitro lymphocyte rosetting
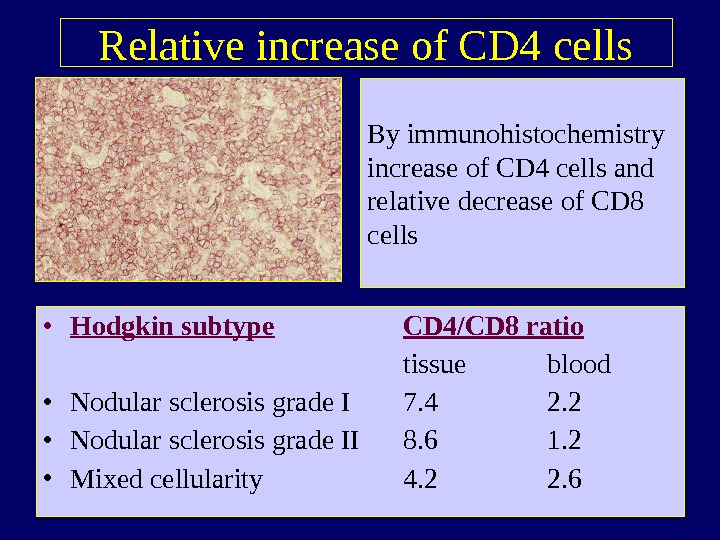
 Relative increase of CD 4 cells • Hodgkin subtype CD 4/CD 8 ratio tissue blood • Nodular sclerosis grade I 7. 4 2. 2 • Nodular sclerosis grade II 8. 6 1. 2 • Mixed cellularity 4. 2 2. 6 By immunohistochemistry increase of CD 4 cells and relative decrease of CD 8 cells
Relative increase of CD 4 cells • Hodgkin subtype CD 4/CD 8 ratio tissue blood • Nodular sclerosis grade I 7. 4 2. 2 • Nodular sclerosis grade II 8. 6 1. 2 • Mixed cellularity 4. 2 2. 6 By immunohistochemistry increase of CD 4 cells and relative decrease of CD 8 cells
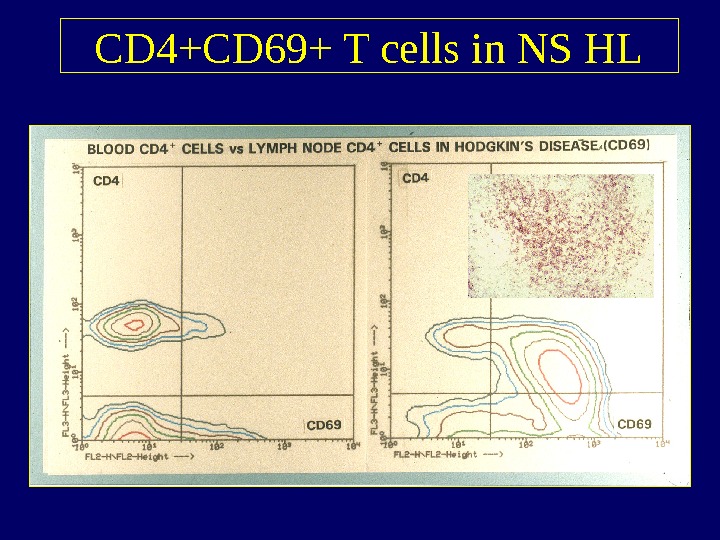
 CD 4+CD 69+ T cells in NS HL
CD 4+CD 69+ T cells in NS HL
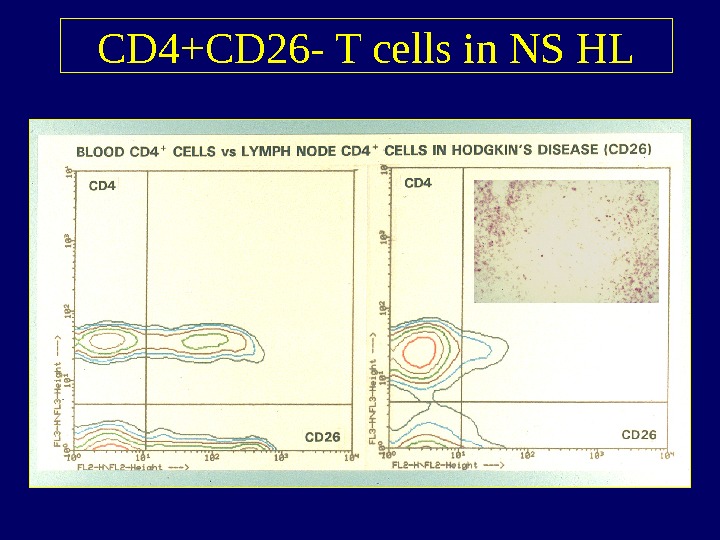
 CD 4+CD 26 — T cells in NS HL
CD 4+CD 26 — T cells in NS HL
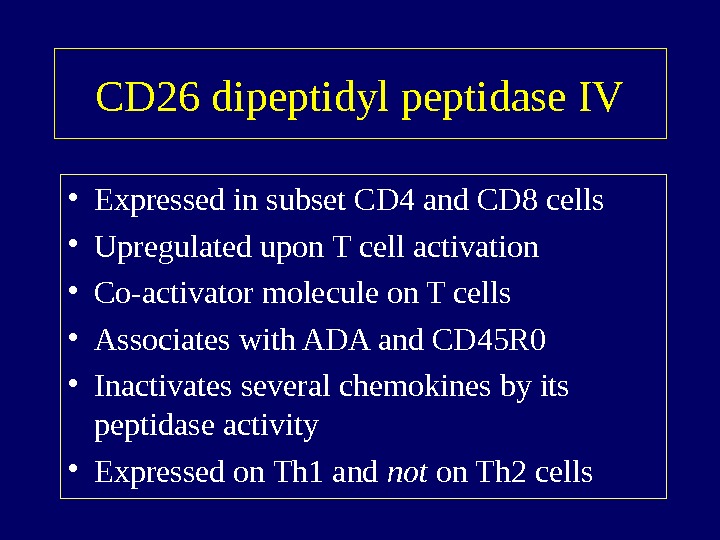
 CD 26 dipeptidyl peptidase IV • Expressed in subset CD 4 and CD 8 cells • Upregulated upon T cell activation • Co-activator molecule on T cells • Associates with ADA and CD 45 R 0 • Inactivates several chemokines by its peptidase activity • Expressed on Th 1 and not on Th 2 cells
CD 26 dipeptidyl peptidase IV • Expressed in subset CD 4 and CD 8 cells • Upregulated upon T cell activation • Co-activator molecule on T cells • Associates with ADA and CD 45 R 0 • Inactivates several chemokines by its peptidase activity • Expressed on Th 1 and not on Th 2 cells
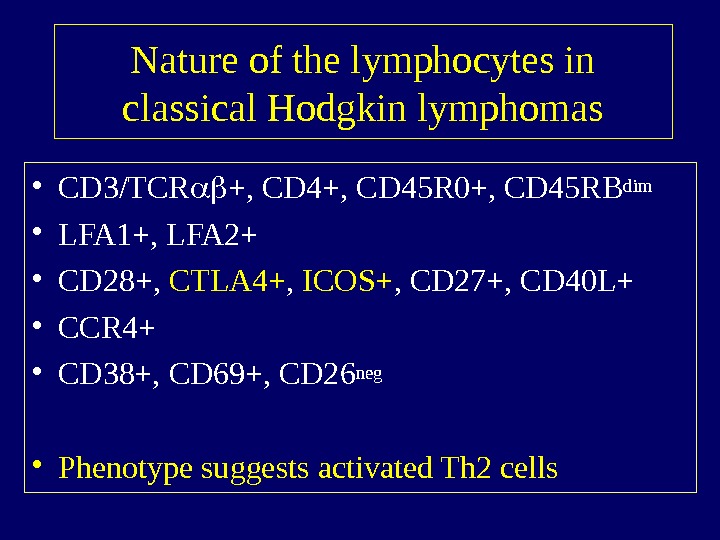
 Nature of the lymphocytes in classical Hodgkin lymphomas • CD 3/TCR +, CD 45 R 0+, CD 45 RBdim • LFA 1+, LFA 2+ • CD 28+, CTLA 4+ , ICOS+ , CD 27+, CD 40 L+ • CCR 4+ • CD 38+, CD 69+, CD 26 neg • Phenotype suggests activated Th 2 cells
Nature of the lymphocytes in classical Hodgkin lymphomas • CD 3/TCR +, CD 45 R 0+, CD 45 RBdim • LFA 1+, LFA 2+ • CD 28+, CTLA 4+ , ICOS+ , CD 27+, CD 40 L+ • CCR 4+ • CD 38+, CD 69+, CD 26 neg • Phenotype suggests activated Th 2 cells
 Co-stimulation • Co-stimulation via CTLA-4 may inhibit T-cell activation. • Co-stimulation via ICOS may lead to TH 2 type cytokine production
Co-stimulation • Co-stimulation via CTLA-4 may inhibit T-cell activation. • Co-stimulation via ICOS may lead to TH 2 type cytokine production
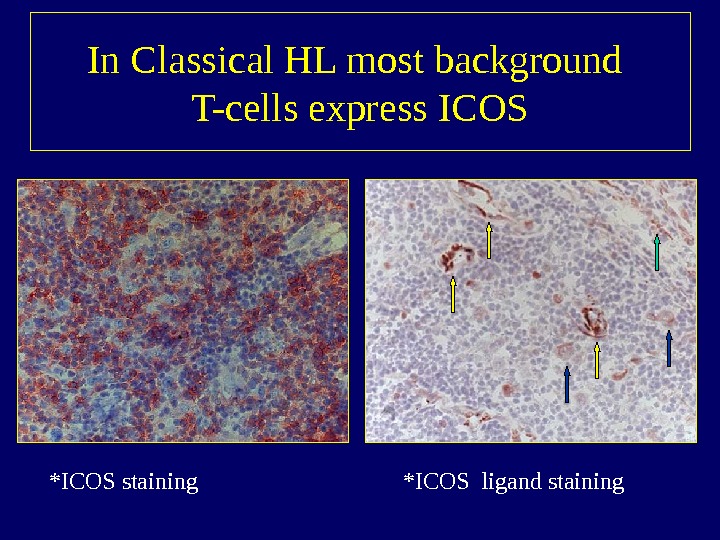
 In Classical HL most background T-cells express ICOS *ICOS staining *ICOS ligand staining
In Classical HL most background T-cells express ICOS *ICOS staining *ICOS ligand staining
 Cytokine production (pg/ml) by Ho LN cells (PMA/ionomycin activation) 02004006008001000 Ho 31 Ho 32 Ho 28 Ho 31 848 717 Ho 32 662 49 Ho 28 611 119 IFNg IL 4 Th 1 Th
Cytokine production (pg/ml) by Ho LN cells (PMA/ionomycin activation) 02004006008001000 Ho 31 Ho 32 Ho 28 Ho 31 848 717 Ho 32 662 49 Ho 28 611 119 IFNg IL 4 Th 1 Th
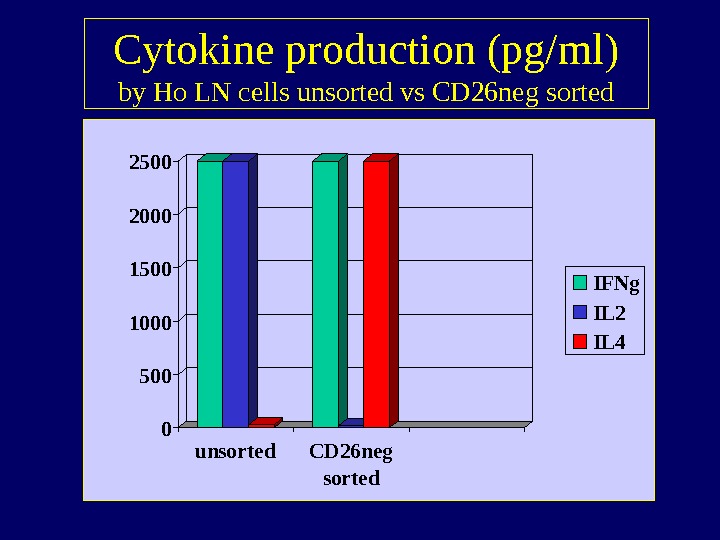
 Cytokine production (pg/ml) by Ho LN cells unsorted vs CD 26 neg sorted 05001000150020002500 unsorted CD 26 neg sorted IFNg IL 2 IL
Cytokine production (pg/ml) by Ho LN cells unsorted vs CD 26 neg sorted 05001000150020002500 unsorted CD 26 neg sorted IFNg IL 2 IL
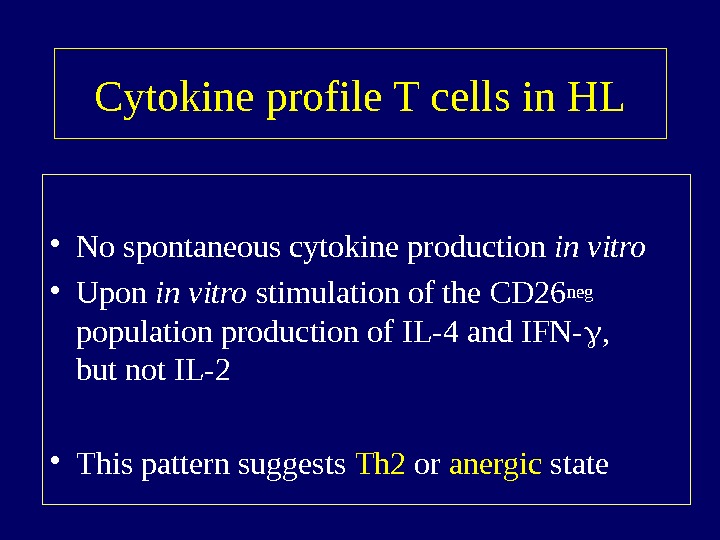
 Cytokine profile T cells in HL • No spontaneous cytokine production in vitro • Upon in vitro stimulation of the CD 26 neg population production of IL-4 and IFN- , but not IL-2 • This pattern suggests Th 2 or anergic state
Cytokine profile T cells in HL • No spontaneous cytokine production in vitro • Upon in vitro stimulation of the CD 26 neg population production of IL-4 and IFN- , but not IL-2 • This pattern suggests Th 2 or anergic state
 Hodgkin Lymphoma & Cytokines/Chemokines C C H Y E T M O O K K I I N N E E S S
Hodgkin Lymphoma & Cytokines/Chemokines C C H Y E T M O O K K I I N N E E S S
 Cytokines and cytokine receptors expressed by R-S cells • IL-1, IL-3, IL-5, IL-6, IL-7, IL-9, IL-10 , IL 13, LT , TGF • IL-2 r , IL-5 r, IL-6 r, IL 9 r, IL-13 r
Cytokines and cytokine receptors expressed by R-S cells • IL-1, IL-3, IL-5, IL-6, IL-7, IL-9, IL-10 , IL 13, LT , TGF • IL-2 r , IL-5 r, IL-6 r, IL 9 r, IL-13 r
 IL 5 receptor in L 428 cells
IL 5 receptor in L 428 cells
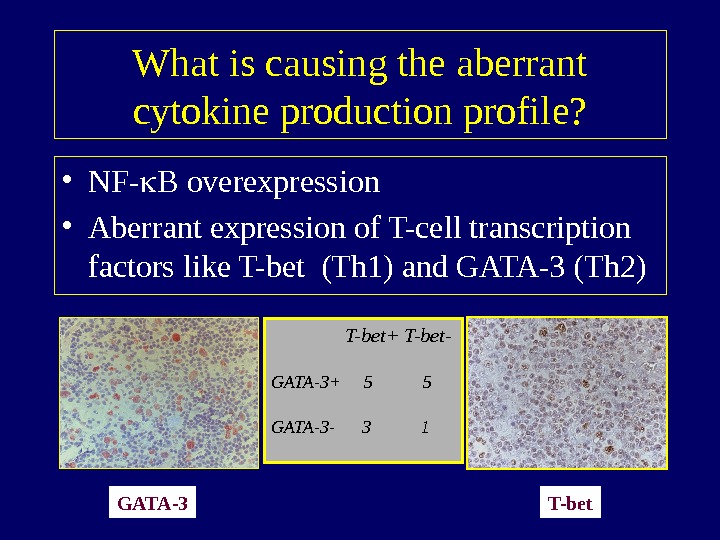
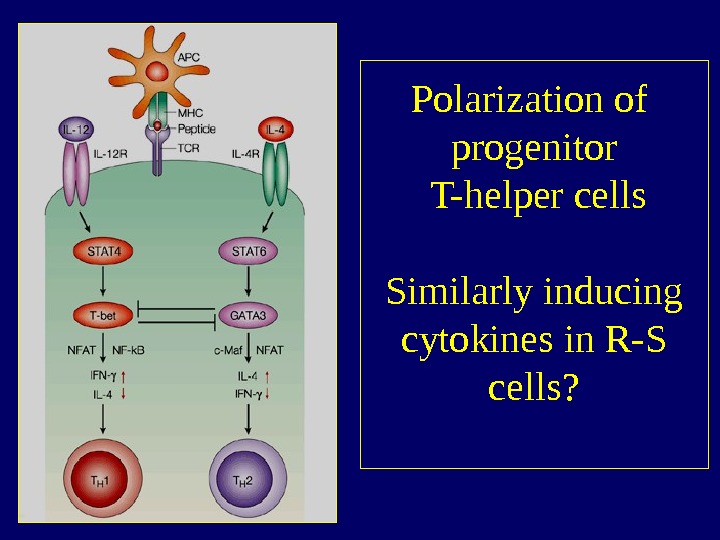
 What is causing the aberrant cytokine production profile? • NF- B overexpression • Aberrant expression of T-cell transcription factors like T-bet (Th 1) and GATA-3 (Th 2) GATA-3 T-bet T-bet+ T-bet- GATA-3+ 5 5 GATA-3 —
What is causing the aberrant cytokine production profile? • NF- B overexpression • Aberrant expression of T-cell transcription factors like T-bet (Th 1) and GATA-3 (Th 2) GATA-3 T-bet T-bet+ T-bet- GATA-3+ 5 5 GATA-3 —
 Polarization of progenitor T-helper cells Similarly inducing cytokines in R-S cells?
Polarization of progenitor T-helper cells Similarly inducing cytokines in R-S cells?
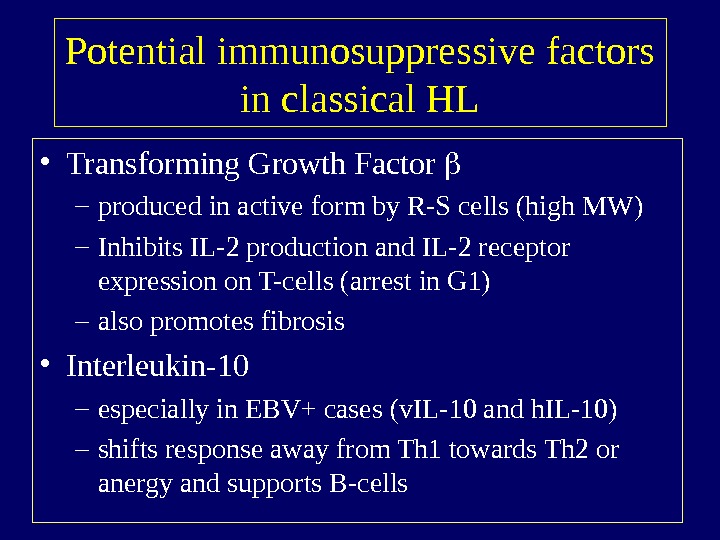
 Potential immunosuppressive factors in classical HL • Transforming Growth Factor – produced in active form by R-S cells (high MW) – Inhibits IL-2 production and IL-2 receptor expression on T-cells (arrest in G 1) – also promotes fibrosis • Interleukin-10 – especially in EBV+ cases (v. IL-10 and h. IL-10) – shifts response away from Th 1 towards Th 2 or anergy and supports B-cells
Potential immunosuppressive factors in classical HL • Transforming Growth Factor – produced in active form by R-S cells (high MW) – Inhibits IL-2 production and IL-2 receptor expression on T-cells (arrest in G 1) – also promotes fibrosis • Interleukin-10 – especially in EBV+ cases (v. IL-10 and h. IL-10) – shifts response away from Th 1 towards Th 2 or anergy and supports B-cells
 TGF ISH in NS HL
TGF ISH in NS HL
 Steps in development of the characteristic infiltrate of HL • Rosetting around R-S cells • Activation of the T lymphocytes • Suppression/immunomodulation • Attraction of lymphocytes into the tissue
Steps in development of the characteristic infiltrate of HL • Rosetting around R-S cells • Activation of the T lymphocytes • Suppression/immunomodulation • Attraction of lymphocytes into the tissue
 Gene expression studies in HL derived cell lines • To identify genes involved in the malignant transformation of R-S precursor cell • To identify genes responsible for the characteristic phenotype of Hodgkin lymphoma • To obtain complete expression profile of classical and L&H type R-S cells (10, 000 tags) • Employing the SAGE technique
Gene expression studies in HL derived cell lines • To identify genes involved in the malignant transformation of R-S precursor cell • To identify genes responsible for the characteristic phenotype of Hodgkin lymphoma • To obtain complete expression profile of classical and L&H type R-S cells (10, 000 tags) • Employing the SAGE technique
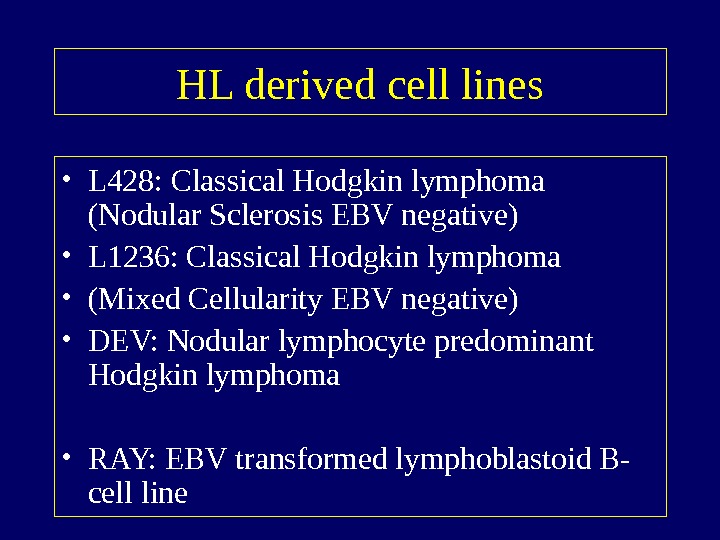
 HL derived cell lines • L 428: Classical Hodgkin lymphoma (Nodular Sclerosis EBV negative) • L 1236: Classical Hodgkin lymphoma • (Mixed Cellularity EBV negative) • DEV: Nodular lymphocyte predominant Hodgkin lymphoma • RAY: EBV transformed lymphoblastoid B- cell line
HL derived cell lines • L 428: Classical Hodgkin lymphoma (Nodular Sclerosis EBV negative) • L 1236: Classical Hodgkin lymphoma • (Mixed Cellularity EBV negative) • DEV: Nodular lymphocyte predominant Hodgkin lymphoma • RAY: EBV transformed lymphoblastoid B- cell line
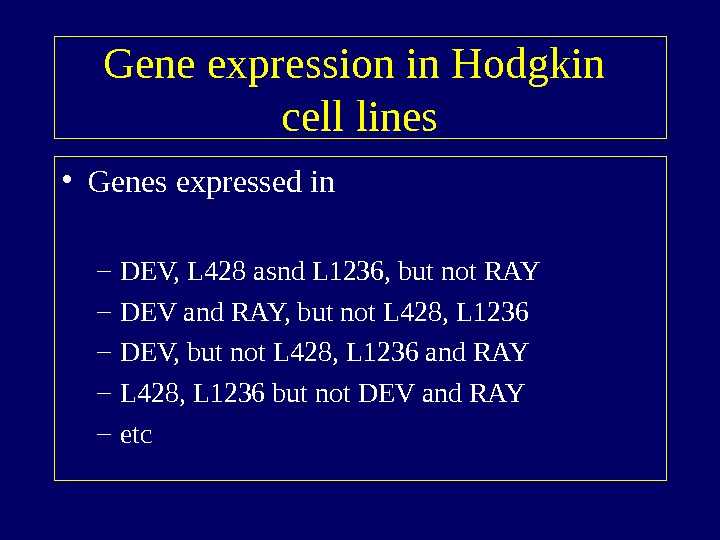
 Gene expression in Hodgkin cell lines • Genes expressed in – DEV, L 428 asnd L 1236, but not RAY – DEV and RAY, but not L 428, L 1236 – DEV, but not L 428, L 1236 and RAY – L 428, L 1236 but not DEV and RAY – etc
Gene expression in Hodgkin cell lines • Genes expressed in – DEV, L 428 asnd L 1236, but not RAY – DEV and RAY, but not L 428, L 1236 – DEV, but not L 428, L 1236 and RAY – L 428, L 1236 but not DEV and RAY – etc
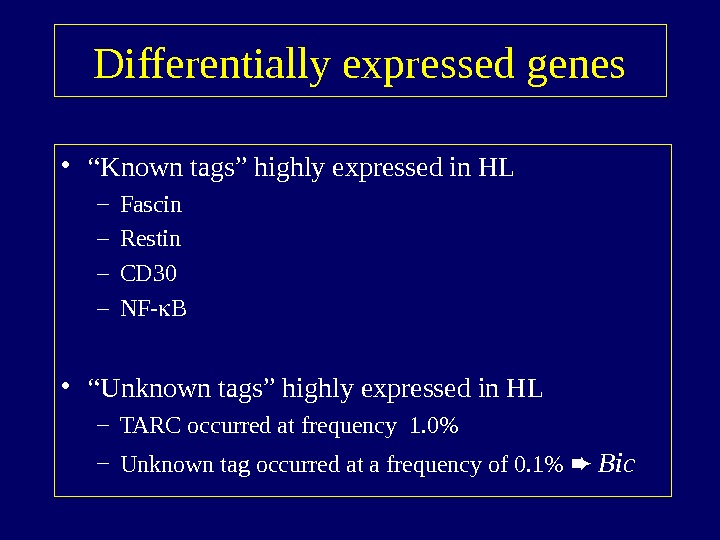
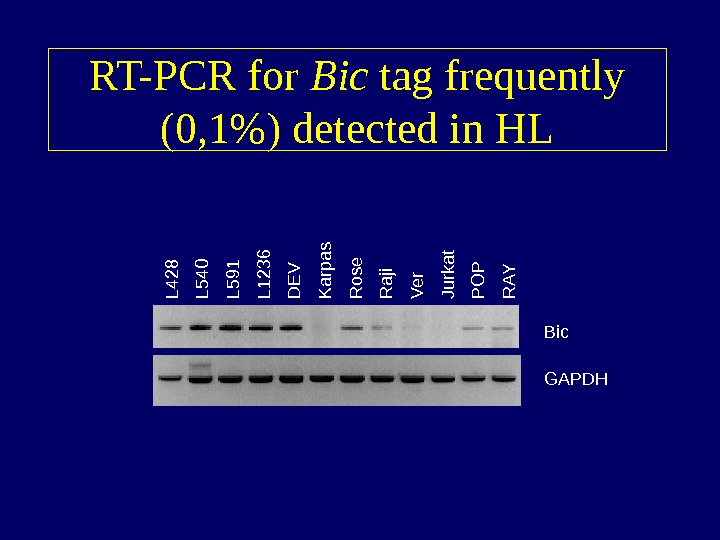
 Differentially expressed genes • “ Known tags” highly expressed in HL – Fascin – Restin – CD 30 – NF- B • “ Unknown tags” highly expressed in HL – TARC occurred at frequency 1. 0% – Unknown tag occurred at a frequency of 0. 1% Bic
Differentially expressed genes • “ Known tags” highly expressed in HL – Fascin – Restin – CD 30 – NF- B • “ Unknown tags” highly expressed in HL – TARC occurred at frequency 1. 0% – Unknown tag occurred at a frequency of 0. 1% Bic
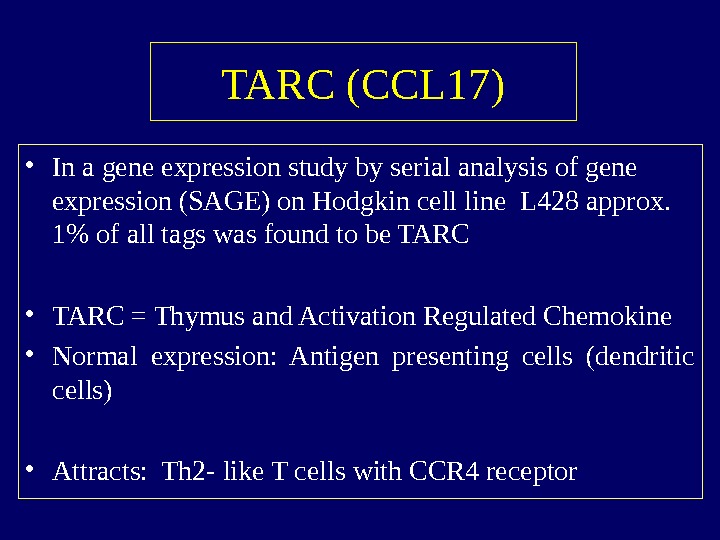
 TARC (CCL 17) • In a gene expression study by serial analysis of gene expression (SAGE) on Hodgkin cell line L 428 approx. 1% of all tags was found to be TARC • TARC = Thymus and Activation Regulated Chemokine • Normal expression: Antigen presenting cells (dendritic cells) • Attracts: Th 2 — like T cells with CCR 4 receptor
TARC (CCL 17) • In a gene expression study by serial analysis of gene expression (SAGE) on Hodgkin cell line L 428 approx. 1% of all tags was found to be TARC • TARC = Thymus and Activation Regulated Chemokine • Normal expression: Antigen presenting cells (dendritic cells) • Attracts: Th 2 — like T cells with CCR 4 receptor
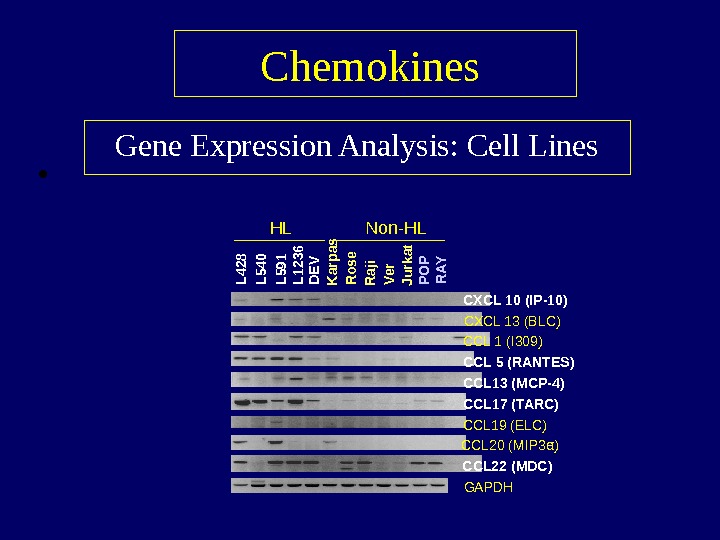
 • Chemokines Gene Expression Analysis: Cell Lines α )CXCL 10 (IP-10) CXCL 13 (BLC) CCL 1 (I 309) CCL 5 (RANTES) CCL 13 (MCP-4) CCL 17 (TARC) CCL 19 (ELC) CCL 20 (MIP 3 a) CCL 22 (MDC) GAPDHL 428 L 540 L 591 L 1236 D EV K arpas R ose R aji Ver Jurkat PO P R AYHL Non-HL
• Chemokines Gene Expression Analysis: Cell Lines α )CXCL 10 (IP-10) CXCL 13 (BLC) CCL 1 (I 309) CCL 5 (RANTES) CCL 13 (MCP-4) CCL 17 (TARC) CCL 19 (ELC) CCL 20 (MIP 3 a) CCL 22 (MDC) GAPDHL 428 L 540 L 591 L 1236 D EV K arpas R ose R aji Ver Jurkat PO P R AYHL Non-HL
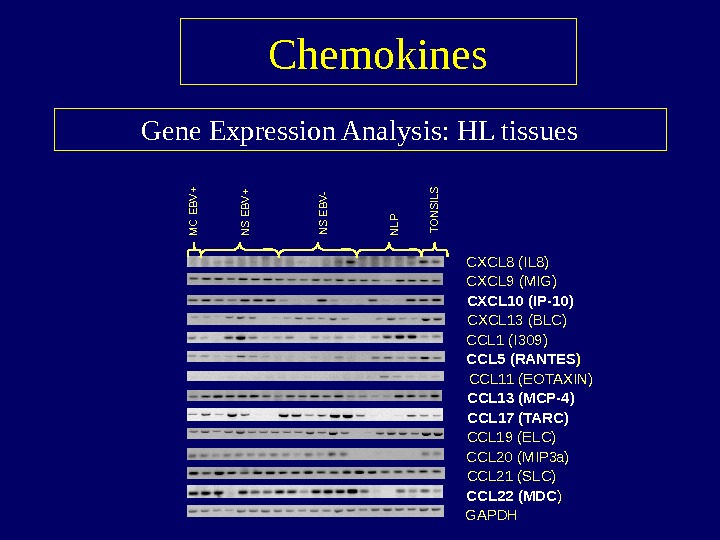
 Gene Expression Analysis: HL tissues Chemokines CXCL 8 (IL 8) CXCL 9 (MIG) CXCL 10 (IP-10) CXCL 13 (BLC) CCL 1 (I 309) CCL 5 (RANTES ) CCL 11 (EOTAXIN) CCL 13 (MCP-4) CCL 17 (TARC) CCL 19 (ELC) CCL 20 (MIP 3 a ) CCL 21 (SLC) CCL 22 (MDC ) GAPDHMC EBV+ NS EBV- NLP TONSILS
Gene Expression Analysis: HL tissues Chemokines CXCL 8 (IL 8) CXCL 9 (MIG) CXCL 10 (IP-10) CXCL 13 (BLC) CCL 1 (I 309) CCL 5 (RANTES ) CCL 11 (EOTAXIN) CCL 13 (MCP-4) CCL 17 (TARC) CCL 19 (ELC) CCL 20 (MIP 3 a ) CCL 21 (SLC) CCL 22 (MDC ) GAPDHMC EBV+ NS EBV- NLP TONSILS
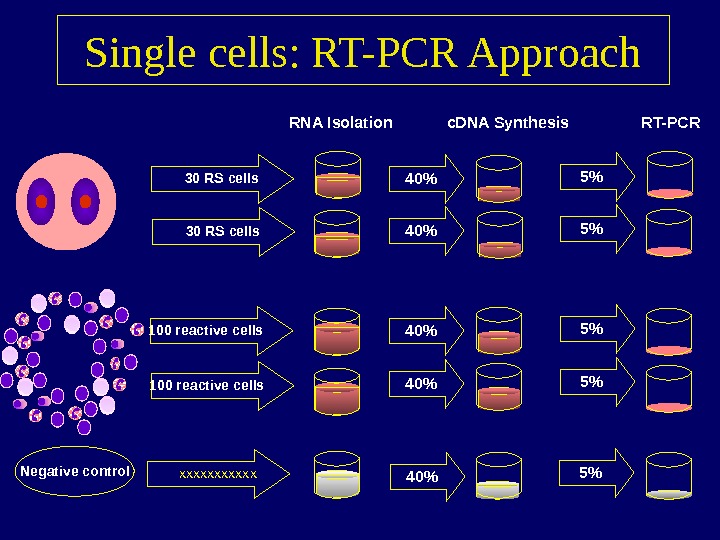
 Single cells: RT-PCR Approach 30 RS cells 100 reactive cells RNA Isolation 40% 40% c. DNA Synthesis 5% 5% RT-PCR Negative control xxxxxx 40% 5%
Single cells: RT-PCR Approach 30 RS cells 100 reactive cells RNA Isolation 40% 40% c. DNA Synthesis 5% 5% RT-PCR Negative control xxxxxx 40% 5%
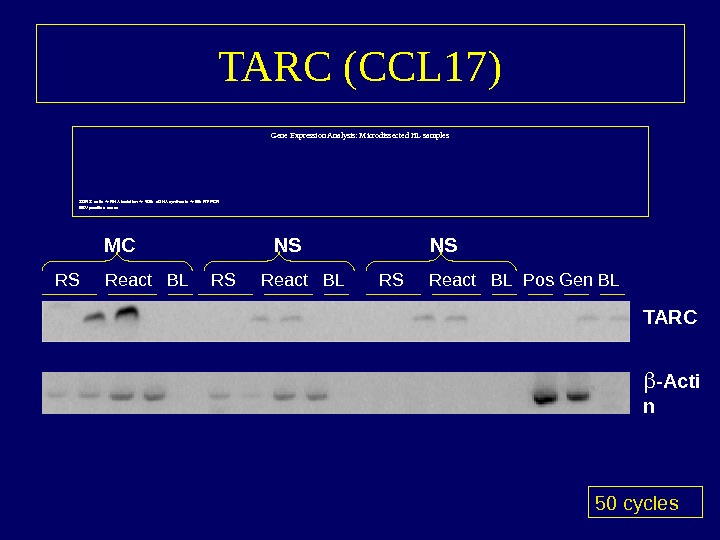
 TARC (CCL 17) Gene Expression Analysis: Microdissected HL samples 30 RS cells RNA isolation 40% c. DNA synthesis 5% RT-PCR EBV positive cases 50 cycles. RS React BL Pos Gen BLMC NS -Acti n. TARCNS RS React BL
TARC (CCL 17) Gene Expression Analysis: Microdissected HL samples 30 RS cells RNA isolation 40% c. DNA synthesis 5% RT-PCR EBV positive cases 50 cycles. RS React BL Pos Gen BLMC NS -Acti n. TARCNS RS React BL
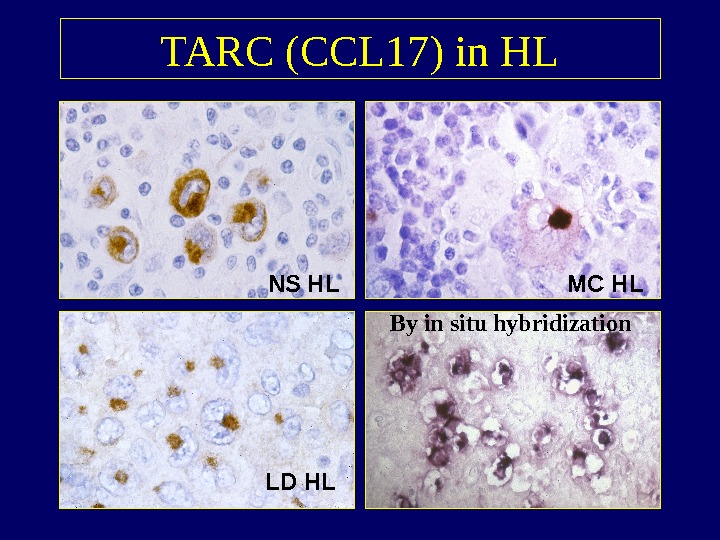
 TARC (CCL 17) in HL NS HL LD HL MC HL By in situ hybridization
TARC (CCL 17) in HL NS HL LD HL MC HL By in situ hybridization
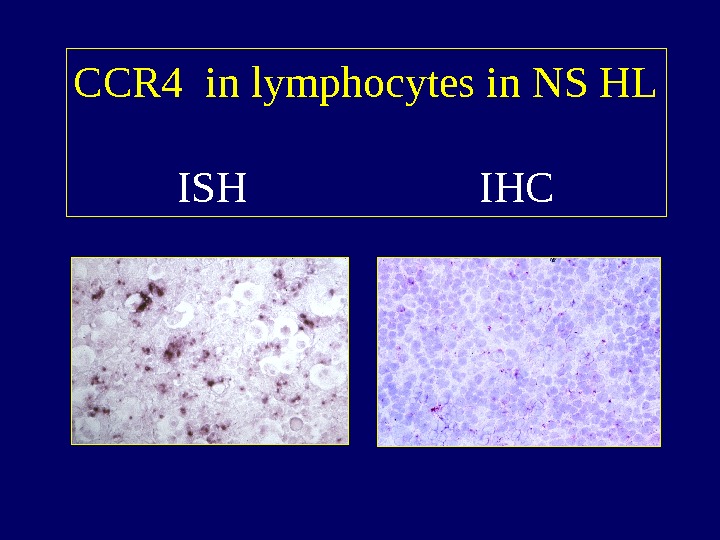
 CCR 4 in lymphocytes in NS HL ISH IH
CCR 4 in lymphocytes in NS HL ISH IH
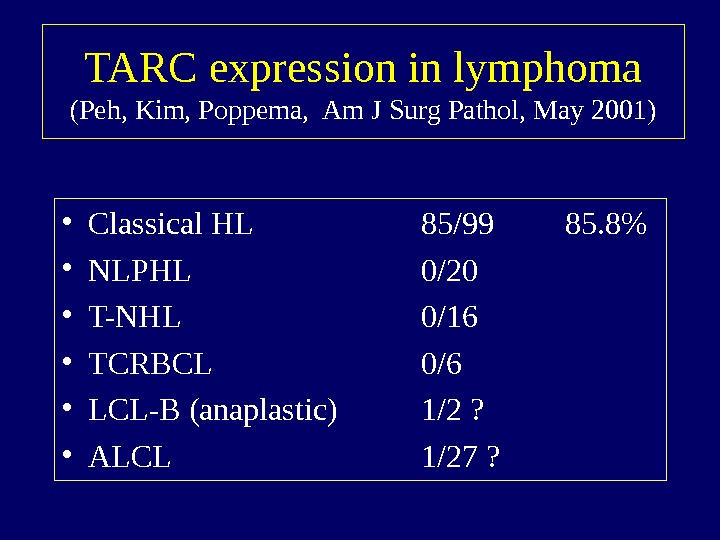
 TARC expression in lymphoma (Peh, Kim, Poppema, Am J Surg Pathol, May 2001) • Classical HL 85/99 85. 8% • NLPHL 0/20 • T-NHL 0/16 • TCRBCL 0/6 • LCL-B (anaplastic) 1/2 ? • ALCL 1/27 ?
TARC expression in lymphoma (Peh, Kim, Poppema, Am J Surg Pathol, May 2001) • Classical HL 85/99 85. 8% • NLPHL 0/20 • T-NHL 0/16 • TCRBCL 0/6 • LCL-B (anaplastic) 1/2 ? • ALCL 1/27 ?
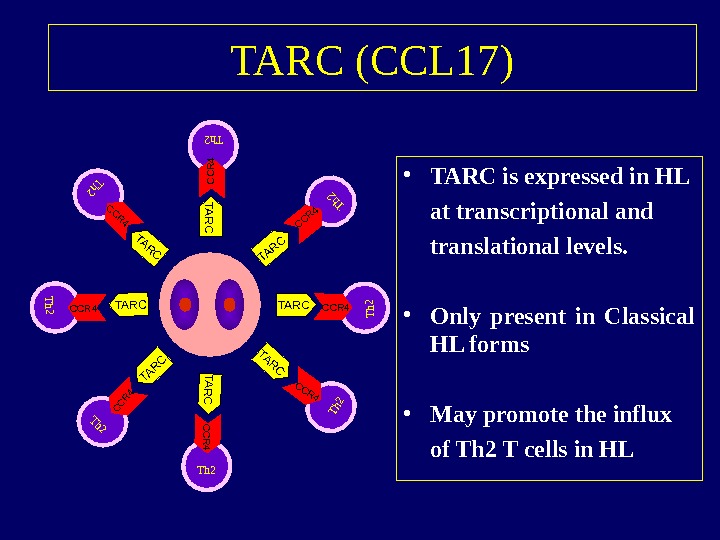
 TARC (CCL 17) • TARC is expressed in HL at transcriptional and translational levels. • Only present in Classical HL forms • May promote the influx of Th 2 T cells in HLTh 2 CCR 4 TARC TARC Th 2 CCR 4 Th 2 CCR 4 Th 2 CCR
TARC (CCL 17) • TARC is expressed in HL at transcriptional and translational levels. • Only present in Classical HL forms • May promote the influx of Th 2 T cells in HLTh 2 CCR 4 TARC TARC Th 2 CCR 4 Th 2 CCR 4 Th 2 CCR
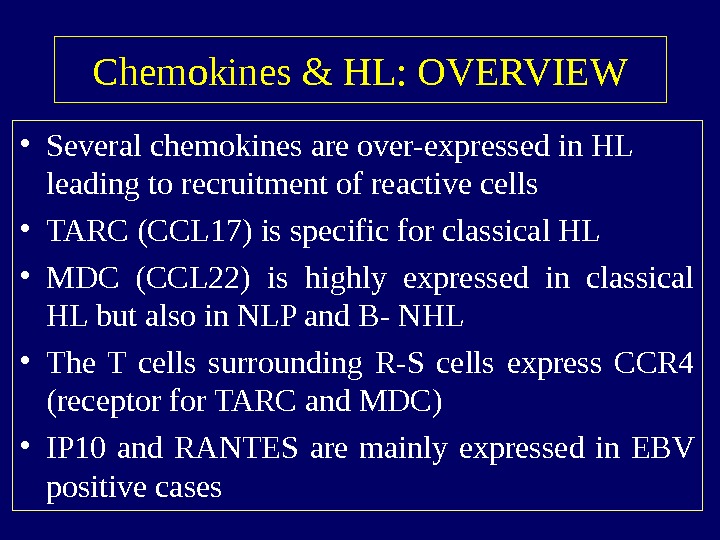
 Chemokines & HL: OVERVIEW • Several chemokines are over-expressed in HL leading to recruitment of reactive cells • TARC (CCL 17) is specific for classical HL • MDC (CCL 22) is highly expressed in classical HL but also in NLP and B- NHL • The T cells surrounding R-S cells express CCR 4 (receptor for TARC and MDC) • IP 10 and RANTES are mainly expressed in EBV positive cases
Chemokines & HL: OVERVIEW • Several chemokines are over-expressed in HL leading to recruitment of reactive cells • TARC (CCL 17) is specific for classical HL • MDC (CCL 22) is highly expressed in classical HL but also in NLP and B- NHL • The T cells surrounding R-S cells express CCR 4 (receptor for TARC and MDC) • IP 10 and RANTES are mainly expressed in EBV positive cases
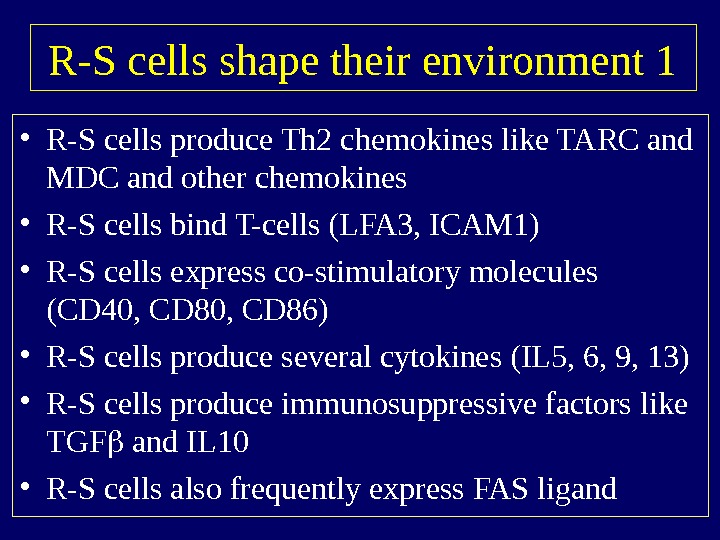
 R-S cells shape their environment 1 • R-S cells produce Th 2 chemokines like TARC and MDC and other chemokines • R-S cells bind T-cells (LFA 3, ICAM 1) • R-S cells express co-stimulatory molecules (CD 40, CD 86) • R-S cells produce several cytokines (IL 5, 6, 9, 13) • R-S cells produce immunosuppressive factors like TGF and IL 10 • R-S cells also frequently express FAS ligand
R-S cells shape their environment 1 • R-S cells produce Th 2 chemokines like TARC and MDC and other chemokines • R-S cells bind T-cells (LFA 3, ICAM 1) • R-S cells express co-stimulatory molecules (CD 40, CD 86) • R-S cells produce several cytokines (IL 5, 6, 9, 13) • R-S cells produce immunosuppressive factors like TGF and IL 10 • R-S cells also frequently express FAS ligand
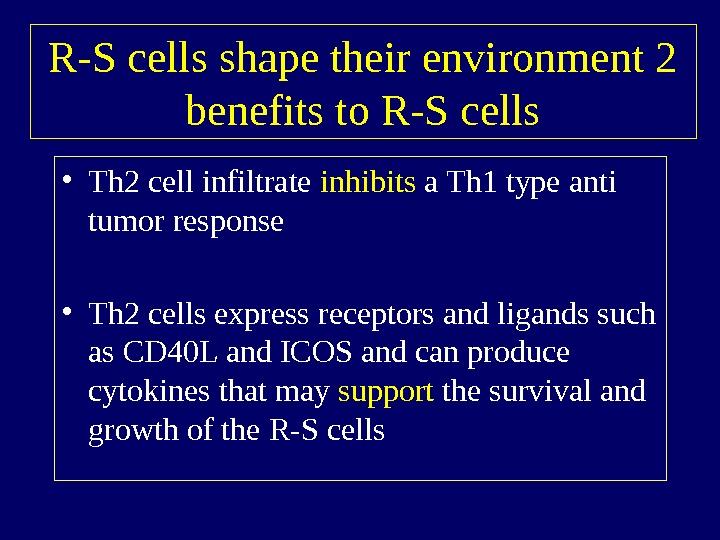
 R-S cells shape their environment 2 benefits to R-S cells • Th 2 cell infiltrate inhibits a Th 1 type anti tumor response • Th 2 cells express receptors and ligands such as CD 40 L and ICOS and can produce cytokines that may support the survival and growth of the R-S cells
R-S cells shape their environment 2 benefits to R-S cells • Th 2 cell infiltrate inhibits a Th 1 type anti tumor response • Th 2 cells express receptors and ligands such as CD 40 L and ICOS and can produce cytokines that may support the survival and growth of the R-S cells
 Bic, a noncoding m. RNA molecule highly expressed in R-S cells • Identified by SAGE in Hodgkin cell lines • Question: • Is this gene of relevance?
Bic, a noncoding m. RNA molecule highly expressed in R-S cells • Identified by SAGE in Hodgkin cell lines • Question: • Is this gene of relevance?
 RT-PCR for Bic tag frequently (0, 1%) detected in HLL 4 2 8 L 5 40 L 5 91 L 12 36 D E V K arpas R ose R a ji V e r Jurkat P O P R A Y Bic GAPDH
RT-PCR for Bic tag frequently (0, 1%) detected in HLL 4 2 8 L 5 40 L 5 91 L 12 36 D E V K arpas R ose R a ji V e r Jurkat P O P R A Y Bic GAPDH
 RNA in-situ hybridization Hodgkin lymphoma, anti-sense Bic probe
RNA in-situ hybridization Hodgkin lymphoma, anti-sense Bic probe
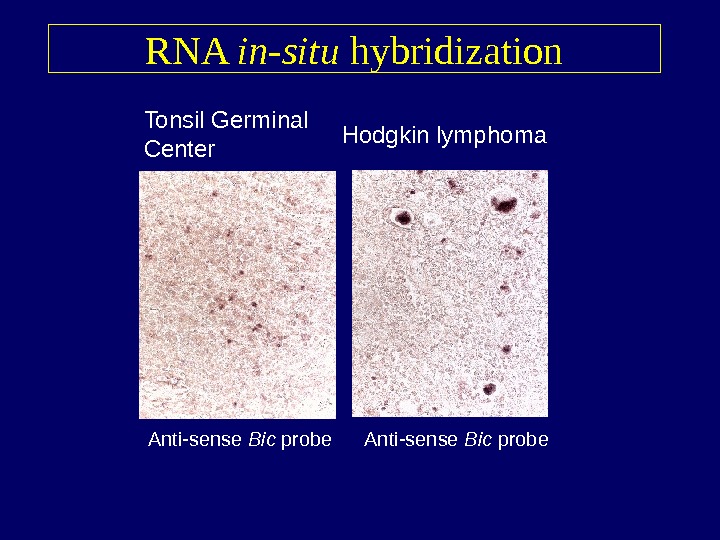
 RNA in-situ hybridization Tonsil Germinal Center Hodgkin lymphoma Anti-sense Bic probe
RNA in-situ hybridization Tonsil Germinal Center Hodgkin lymphoma Anti-sense Bic probe
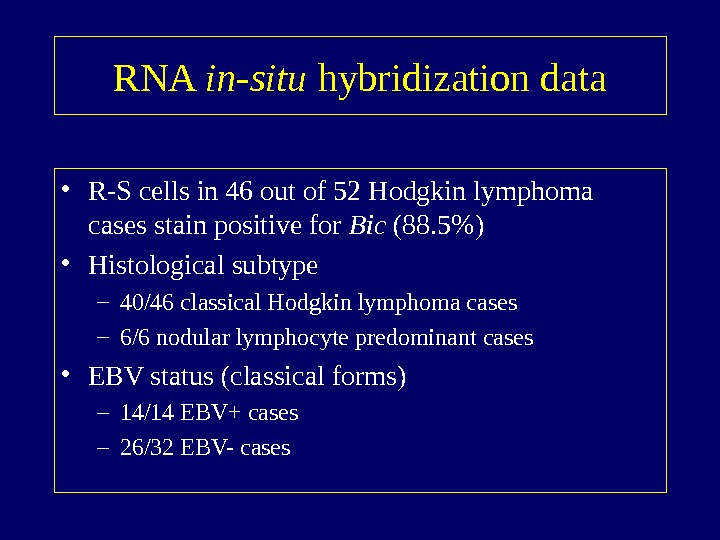
 RNA in-situ hybridization data • R-S cells in 46 out of 52 Hodgkin lymphoma cases stain positive for Bic (88. 5%) • Histological subtype – 40/46 classical Hodgkin lymphoma cases – 6/6 nodular lymphocyte predominant cases • EBV status (classical forms) – 14/14 EBV+ cases – 26/32 EBV- cases
RNA in-situ hybridization data • R-S cells in 46 out of 52 Hodgkin lymphoma cases stain positive for Bic (88. 5%) • Histological subtype – 40/46 classical Hodgkin lymphoma cases – 6/6 nodular lymphocyte predominant cases • EBV status (classical forms) – 14/14 EBV+ cases – 26/32 EBV- cases
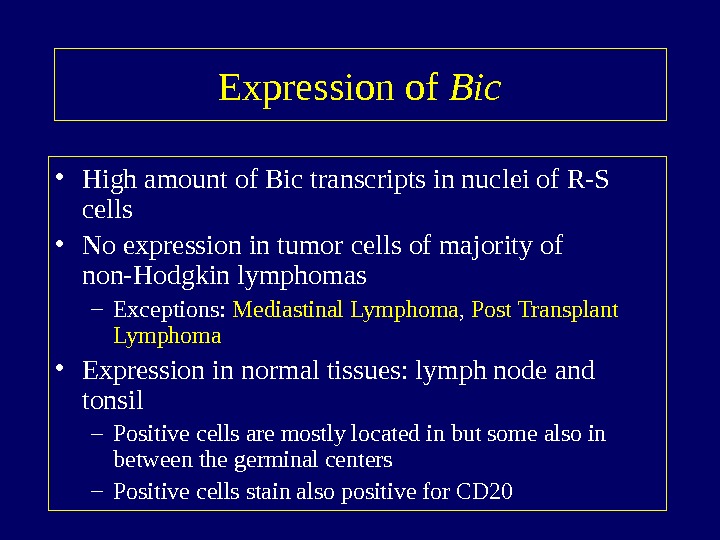
 Expression of Bic • High amount of Bic transcripts in nuclei of R-S cells • No expression in tumor cells of majority of non-Hodgkin lymphomas – Exceptions: Mediastinal Lymphoma , Post Transplant Lymphoma • Expression in normal tissues: lymph node and tonsil – Positive cells are mostly located in but some also in between the germinal centers – Positive cells stain also positive for
Expression of Bic • High amount of Bic transcripts in nuclei of R-S cells • No expression in tumor cells of majority of non-Hodgkin lymphomas – Exceptions: Mediastinal Lymphoma , Post Transplant Lymphoma • Expression in normal tissues: lymph node and tonsil – Positive cells are mostly located in but some also in between the germinal centers – Positive cells stain also positive for
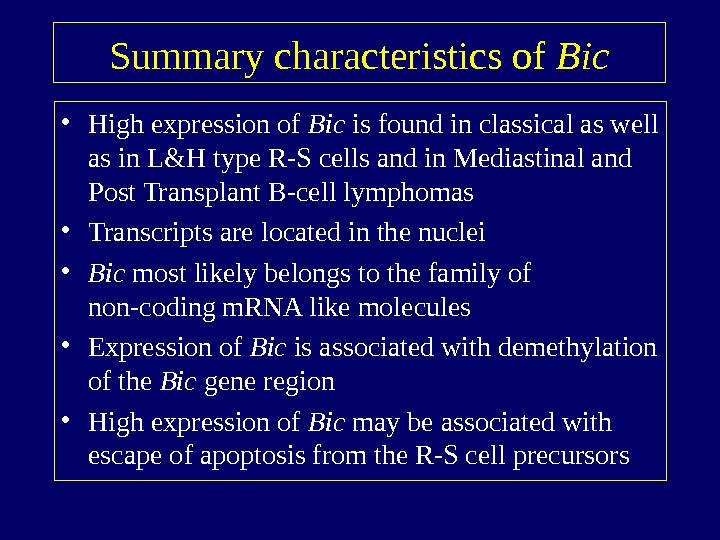
 Summary characteristics of Bic • High expression of Bic is found in classical as well as in L&H type R-S cells and in Mediastinal and Post Transplant B-cell lymphomas • Transcripts are located in the nuclei • Bic most likely belongs to the family of non-coding m. RNA like molecules • Expression of Bic is associated with demethylation of the Bic gene region • High expression of Bic may be associated with escape of apoptosis from the R-S cell precursors
Summary characteristics of Bic • High expression of Bic is found in classical as well as in L&H type R-S cells and in Mediastinal and Post Transplant B-cell lymphomas • Transcripts are located in the nuclei • Bic most likely belongs to the family of non-coding m. RNA like molecules • Expression of Bic is associated with demethylation of the Bic gene region • High expression of Bic may be associated with escape of apoptosis from the R-S cell precursors
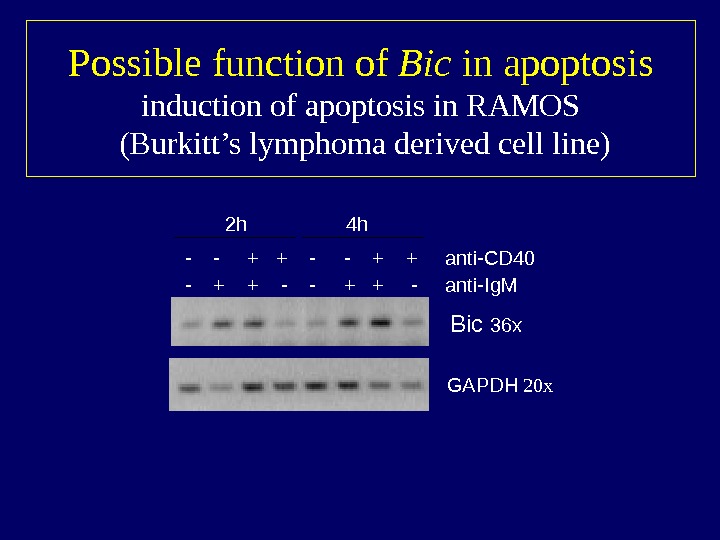
 Possible function of Bic in apoptosis induction of apoptosis in RAMOS (Burkitt’s lymphoma derived cell line) Bic 36 x GAPDH 20 x- — + + — — + + anti-CD 40 — + + — — + + — anti-Ig. M 2 h 4 h
Possible function of Bic in apoptosis induction of apoptosis in RAMOS (Burkitt’s lymphoma derived cell line) Bic 36 x GAPDH 20 x- — + + — — + + anti-CD 40 — + + — — + + — anti-Ig. M 2 h 4 h
 What is the goal? • To take advantage of the weak points of the R-S cells! – Dependence on Th 2 cells – Inherent apoptotic nature • But how to test?
What is the goal? • To take advantage of the weak points of the R-S cells! – Dependence on Th 2 cells – Inherent apoptotic nature • But how to test?
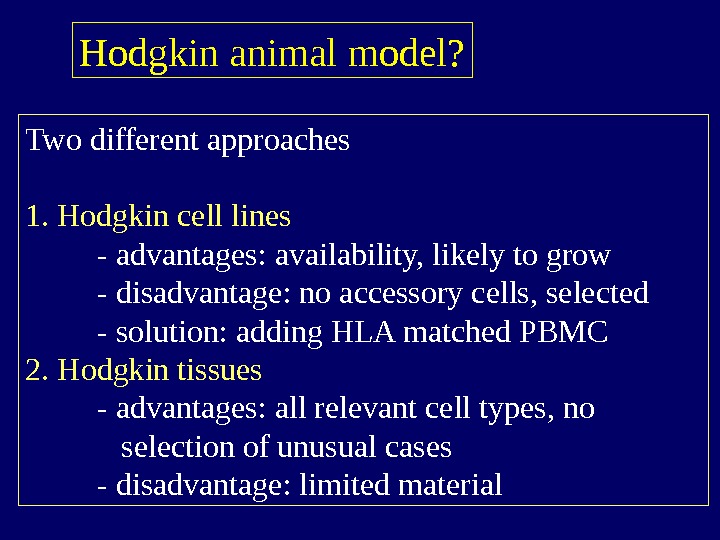
 Hodgkin animal model? Two different approaches 1. Hodgkin cell lines — advantages: availability, likely to grow — disadvantage: no accessory cells, selected — solution: adding HLA matched PBMC 2. Hodgkin tissues — advantages: all relevant cell types, no selection of unusual cases — disadvantage: limited material
Hodgkin animal model? Two different approaches 1. Hodgkin cell lines — advantages: availability, likely to grow — disadvantage: no accessory cells, selected — solution: adding HLA matched PBMC 2. Hodgkin tissues — advantages: all relevant cell types, no selection of unusual cases — disadvantage: limited material
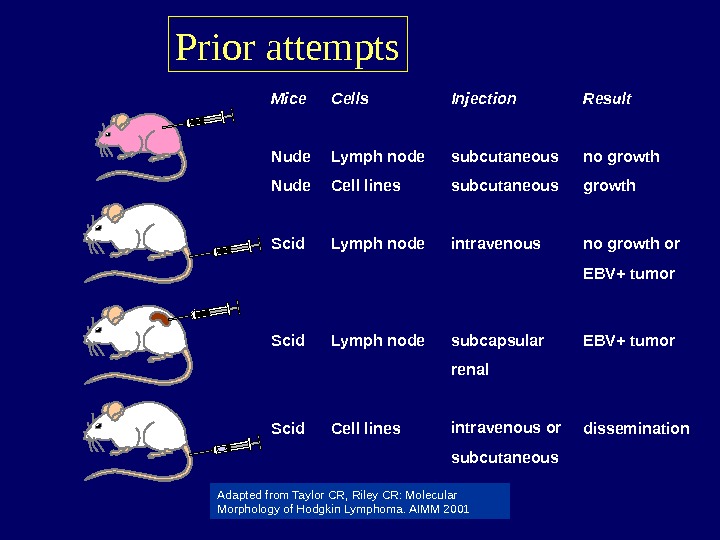
 Prior attempts Mice Nude Scid Cells Lymph node Cell lines Injection subcutaneous intravenous subcapsular renal intravenous or subcutaneous Result no growth or EBV+ tumor dissemination Adapted from Taylor CR, Riley CR: Molecular Morphology of Hodgkin Lymphoma. AIMM
Prior attempts Mice Nude Scid Cells Lymph node Cell lines Injection subcutaneous intravenous subcapsular renal intravenous or subcutaneous Result no growth or EBV+ tumor dissemination Adapted from Taylor CR, Riley CR: Molecular Morphology of Hodgkin Lymphoma. AIMM
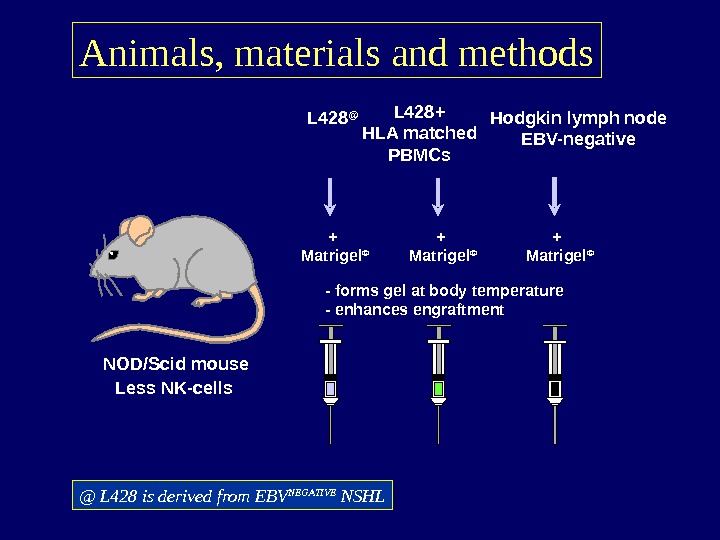
 Animals, materials and methods NOD/Scid mouse Less NK-cells L 428 @ Hodgkin lymph node EBV-negative — forms gel at body temperature — enhances engraftment L 428+ HLA matched PBMCs + Matrigel © @ L 428 is derived from EBV NEGATIVE NSHL
Animals, materials and methods NOD/Scid mouse Less NK-cells L 428 @ Hodgkin lymph node EBV-negative — forms gel at body temperature — enhances engraftment L 428+ HLA matched PBMCs + Matrigel © @ L 428 is derived from EBV NEGATIVE NSHL
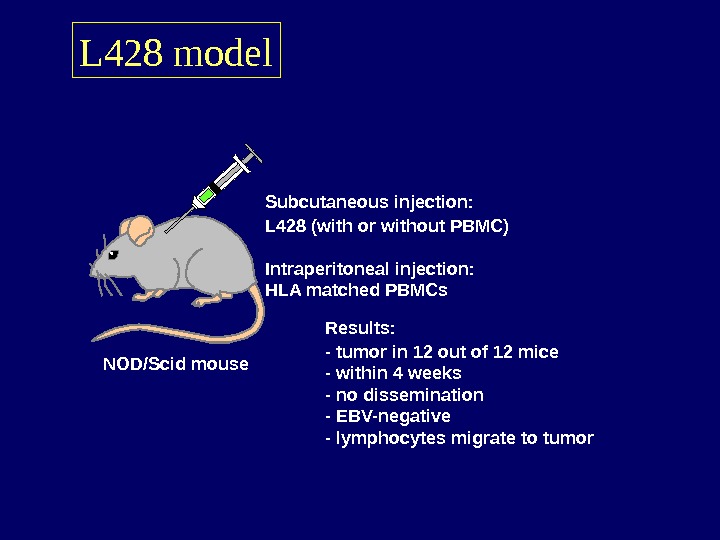
 L 428 model NOD/Scid mouse Subcutaneous injection: L 428 (with or without PBMC) Intraperitoneal injection: HLA matched PBMCs Results: — tumor in 12 out of 12 mice — within 4 weeks — no dissemination — EBV-negative — lymphocytes migrate to tumor
L 428 model NOD/Scid mouse Subcutaneous injection: L 428 (with or without PBMC) Intraperitoneal injection: HLA matched PBMCs Results: — tumor in 12 out of 12 mice — within 4 weeks — no dissemination — EBV-negative — lymphocytes migrate to tumor
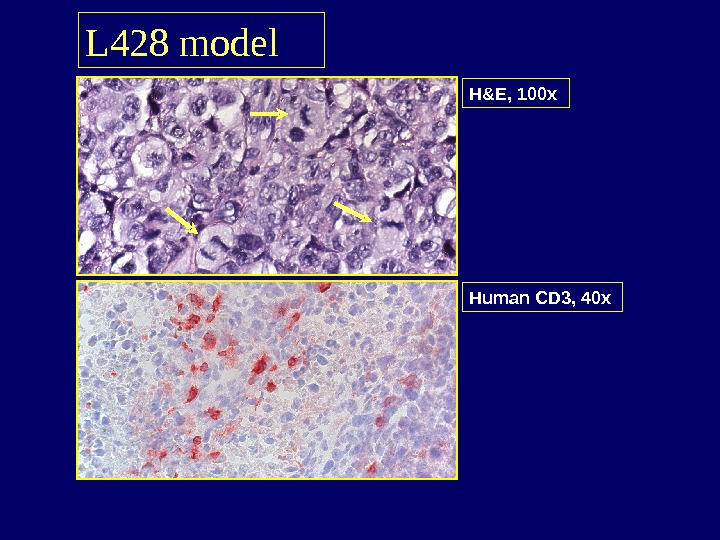
 L 428 model H&E, 100 x Human CD 3, 40 x
L 428 model H&E, 100 x Human CD 3, 40 x
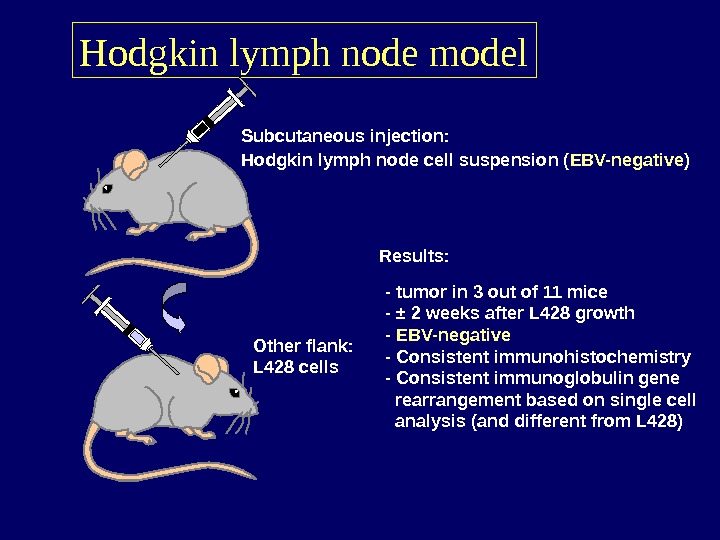
 Hodgkin lymph node model Subcutaneous injection: Hodgkin lymph node cell suspension ( EBV-negative ) Other flank: L 428 cells Results: — tumor in 3 out of 11 mice — ± 2 weeks after L 428 growth — EBV-negative — Consistent immunohistochemistry — Consistent immunoglobulin gene rearrangement based on single cell analysis (and different from L 428)
Hodgkin lymph node model Subcutaneous injection: Hodgkin lymph node cell suspension ( EBV-negative ) Other flank: L 428 cells Results: — tumor in 3 out of 11 mice — ± 2 weeks after L 428 growth — EBV-negative — Consistent immunohistochemistry — Consistent immunoglobulin gene rearrangement based on single cell analysis (and different from L 428)
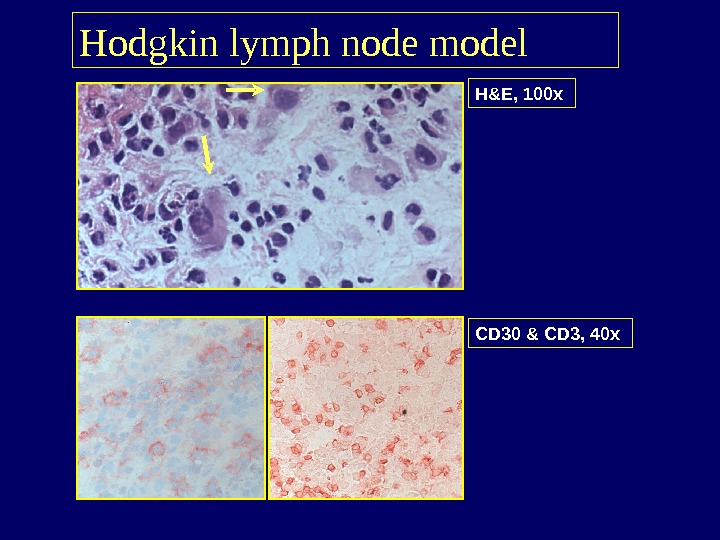
 H&E, 100 x Hodgkin lymph node model CD 30 & CD 3, 40 x
H&E, 100 x Hodgkin lymph node model CD 30 & CD 3, 40 x
 Conclusion / Summary 2 in vivo Hodgkin Lymphoma models: — reproducible — relevant Models can be used for: — studies on interactions between Reed- Sternberg cells and surrounding cells — testing new therapeutic strategies
Conclusion / Summary 2 in vivo Hodgkin Lymphoma models: — reproducible — relevant Models can be used for: — studies on interactions between Reed- Sternberg cells and surrounding cells — testing new therapeutic strategies
 Hodgkin Research Group Department of Pathology & Laboratory Medicine University Hospital Groningen • Debora de Jong, BSc • Geert Harms, BSc • Jane Briggs, BSc • Arjan Diepstra, MD • Joost Kluiver, MSc • Renate Rust, BSc • Cigdem Atayar, MD • Anneke Bosga, BSc • Anke van den Berg, Ph. D • Lydia Visser, Ph. D • Suat Cheng Peh, MD, Ph. D (Kuala Lumpur, Malaysia) • Ewerton Maggio, MD, Ph. D (Curitiba, Brazil)
Hodgkin Research Group Department of Pathology & Laboratory Medicine University Hospital Groningen • Debora de Jong, BSc • Geert Harms, BSc • Jane Briggs, BSc • Arjan Diepstra, MD • Joost Kluiver, MSc • Renate Rust, BSc • Cigdem Atayar, MD • Anneke Bosga, BSc • Anke van den Berg, Ph. D • Lydia Visser, Ph. D • Suat Cheng Peh, MD, Ph. D (Kuala Lumpur, Malaysia) • Ewerton Maggio, MD, Ph. D (Curitiba, Brazil)
