db5bd5297111f2b1e6af9a35af1635bf.ppt
- Количество слайдов: 16
 Segment Alignment (SEA) Yuzhen Ye Adam Godzik The Burnham Institute
Segment Alignment (SEA) Yuzhen Ye Adam Godzik The Burnham Institute
 Outline • • A new look at the local structure prediction Network matching problem Practical issues Applications
Outline • • A new look at the local structure prediction Network matching problem Practical issues Applications
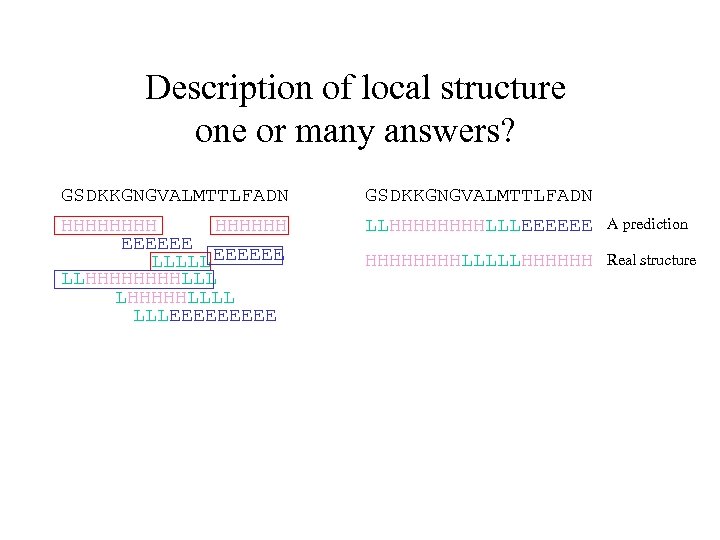 Description of local structure one or many answers? GSDKKGNGVALMTTLFADN HHHHHH EEEEEE LLLLL EEEEEE LLHHHHLLL LHHHHHLLLL LLLEEEEE GSDKKGNGVALMTTLFADN LLHHHHLLLEEEEEE A prediction HHHHLLLLLHHHHHH Real structure
Description of local structure one or many answers? GSDKKGNGVALMTTLFADN HHHHHH EEEEEE LLLLL EEEEEE LLHHHHLLL LHHHHHLLLL LLLEEEEE GSDKKGNGVALMTTLFADN LLHHHHLLLEEEEEE A prediction HHHHLLLLLHHHHHH Real structure
 Motivation • A natural description of local structures: keep the segment information of local structures • Keep uncertainties in local structure predictions: drawbacks of prediction programs and intrinsic uncertainties of local structures in absence of global interactions Incorporating the protein local structure in protein sequence comparison may help to detect the distant homologies and to improve their alignments (for homology modeling)
Motivation • A natural description of local structures: keep the segment information of local structures • Keep uncertainties in local structure predictions: drawbacks of prediction programs and intrinsic uncertainties of local structures in absence of global interactions Incorporating the protein local structure in protein sequence comparison may help to detect the distant homologies and to improve their alignments (for homology modeling)
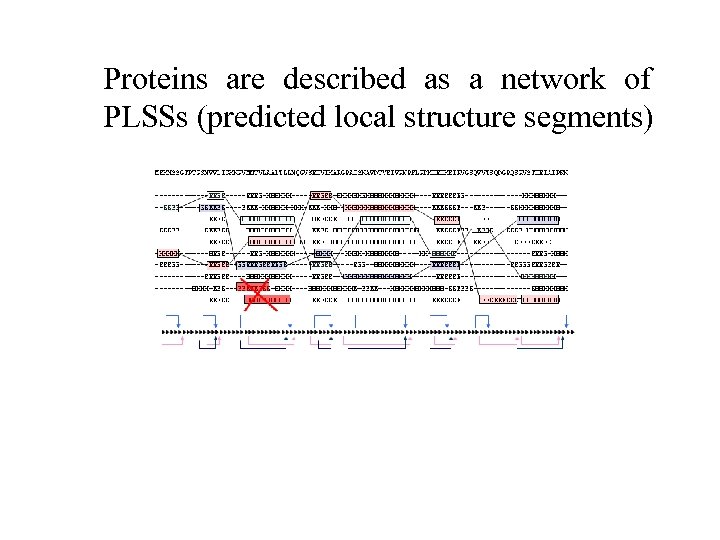 Proteins are described as a network of PLSSs (predicted local structure segments)
Proteins are described as a network of PLSSs (predicted local structure segments)
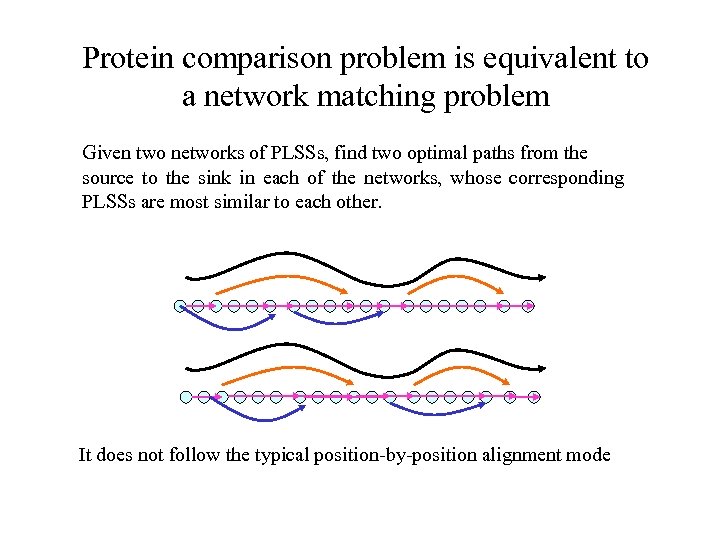 Protein comparison problem is equivalent to a network matching problem Given two networks of PLSSs, find two optimal paths from the source to the sink in each of the networks, whose corresponding PLSSs are most similar to each other. It does not follow the typical position-by-position alignment mode
Protein comparison problem is equivalent to a network matching problem Given two networks of PLSSs, find two optimal paths from the source to the sink in each of the networks, whose corresponding PLSSs are most similar to each other. It does not follow the typical position-by-position alignment mode
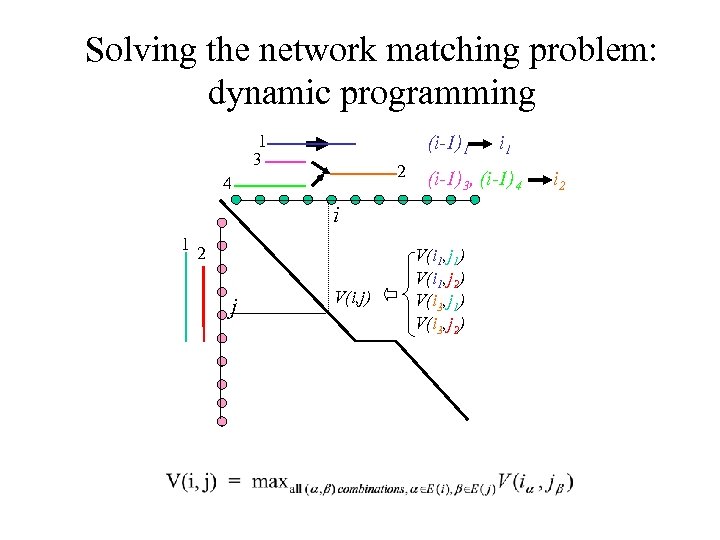 Solving the network matching problem: dynamic programming 1 3 (i-1)1 2 4 (i-1)3, (i-1)4 i 12 j V(i, j) i 1 V(i 1, j 1) V(i 1, j 2) V(i 3, j 1) V(i 3, j 2) i 2
Solving the network matching problem: dynamic programming 1 3 (i-1)1 2 4 (i-1)3, (i-1)4 i 12 j V(i, j) i 1 V(i 1, j 1) V(i 1, j 2) V(i 3, j 1) V(i 3, j 2) i 2
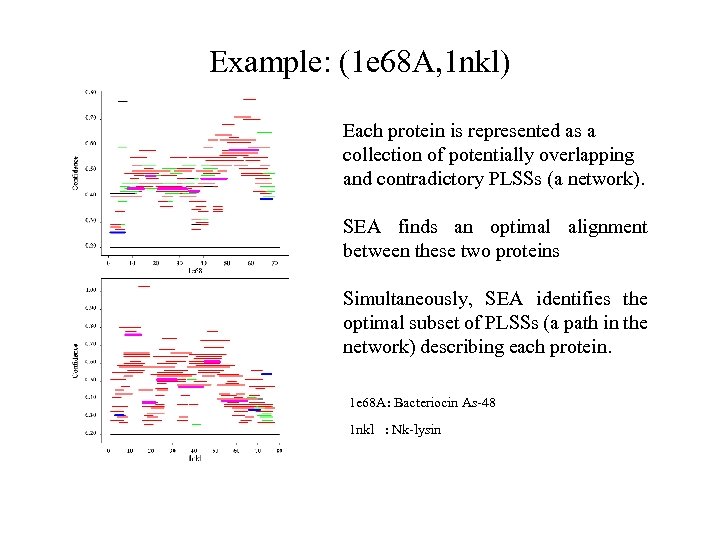 Example: (1 e 68 A, 1 nkl) Each protein is represented as a collection of potentially overlapping and contradictory PLSSs (a network). SEA finds an optimal alignment between these two proteins Simultaneously, SEA identifies the optimal subset of PLSSs (a path in the network) describing each protein. 1 e 68 A: Bacteriocin As-48 1 nkl : Nk-lysin
Example: (1 e 68 A, 1 nkl) Each protein is represented as a collection of potentially overlapping and contradictory PLSSs (a network). SEA finds an optimal alignment between these two proteins Simultaneously, SEA identifies the optimal subset of PLSSs (a path in the network) describing each protein. 1 e 68 A: Bacteriocin As-48 1 nkl : Nk-lysin
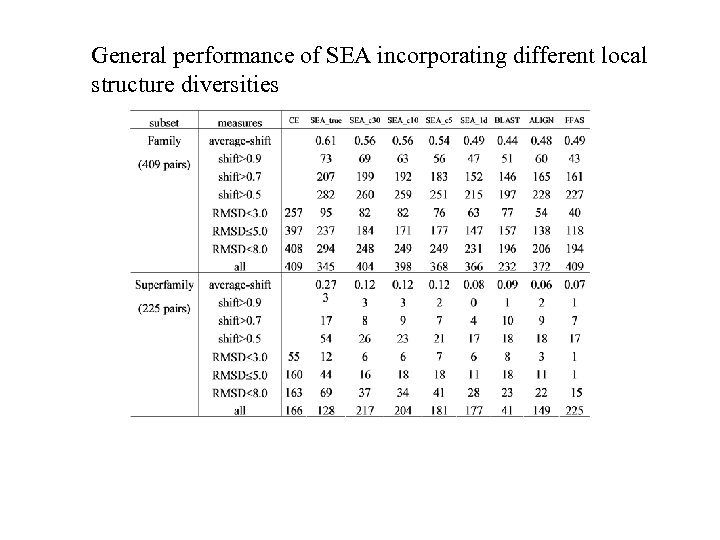 General performance of SEA incorporating different local structure diversities
General performance of SEA incorporating different local structure diversities
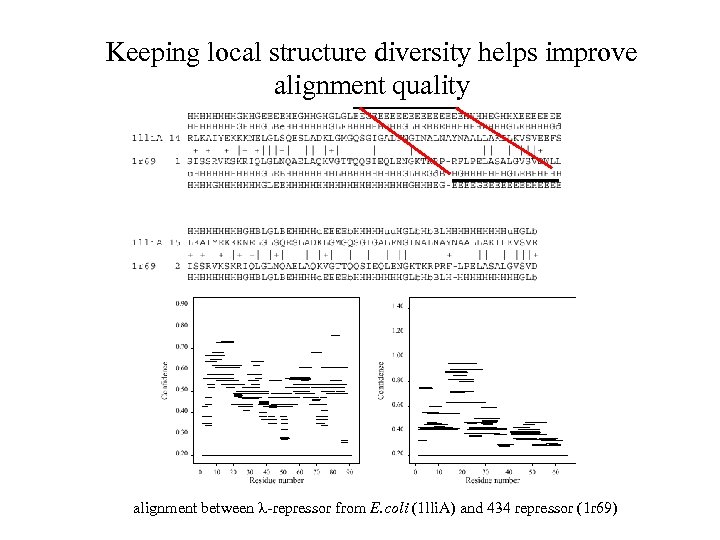 Keeping local structure diversity helps improve alignment quality alignment between -repressor from E. coli (1 lli. A) and 434 repressor (1 r 69)
Keeping local structure diversity helps improve alignment quality alignment between -repressor from E. coli (1 lli. A) and 434 repressor (1 r 69)
 Local structure information is crucial for improving alignments, especially in the more divergent regions Variable region Stable region 1 esf. A: straphylococcal enterotoxin 2 tss. A: toxic shock syndrome toxin-1
Local structure information is crucial for improving alignments, especially in the more divergent regions Variable region Stable region 1 esf. A: straphylococcal enterotoxin 2 tss. A: toxic shock syndrome toxin-1
 Practical issue: local structural prediction • Searching I-site database (web-server or standalone program) • Our solution: Frag. Lib – using sensitive profile-profile alignment program FFAS to predict local structures
Practical issue: local structural prediction • Searching I-site database (web-server or standalone program) • Our solution: Frag. Lib – using sensitive profile-profile alignment program FFAS to predict local structures
 Applications • Distant homology detection • Local structure prediction • Improving alignments for protein modeling
Applications • Distant homology detection • Local structure prediction • Improving alignments for protein modeling
 Reference A segment alignment approach to protein comparison (Bioinformatics, April issue) Web server http: //ffas. ljcrf. edu/sea
Reference A segment alignment approach to protein comparison (Bioinformatics, April issue) Web server http: //ffas. ljcrf. edu/sea
 Related work • Spliced sequence alignment – Gelfand et al. , 1996, PNAS; Novichkov et al. , 2001 – Assembling genes from alternative exons • Jumping alignment – Spang R, Rehmsmeier M, Stoye J. JCB, 2002 – Computes a local alignment of a single sequence and a multiple alignment – The sequence is at each position aligned to one sequence of the multiple alignment (reference sequence) instead of a profile • Partial order alignment – Lee C, Grasso C, Sharlow MF, Bioinformatics, 2002 – Multiple alignment
Related work • Spliced sequence alignment – Gelfand et al. , 1996, PNAS; Novichkov et al. , 2001 – Assembling genes from alternative exons • Jumping alignment – Spang R, Rehmsmeier M, Stoye J. JCB, 2002 – Computes a local alignment of a single sequence and a multiple alignment – The sequence is at each position aligned to one sequence of the multiple alignment (reference sequence) instead of a profile • Partial order alignment – Lee C, Grasso C, Sharlow MF, Bioinformatics, 2002 – Multiple alignment
 Acknowledgements • • Dariusz Plewczyński Iddo Friedberg Łukasz Jaroszewski Weizhong Li • This project is supported by SPAM grant GM 63208
Acknowledgements • • Dariusz Plewczyński Iddo Friedberg Łukasz Jaroszewski Weizhong Li • This project is supported by SPAM grant GM 63208


