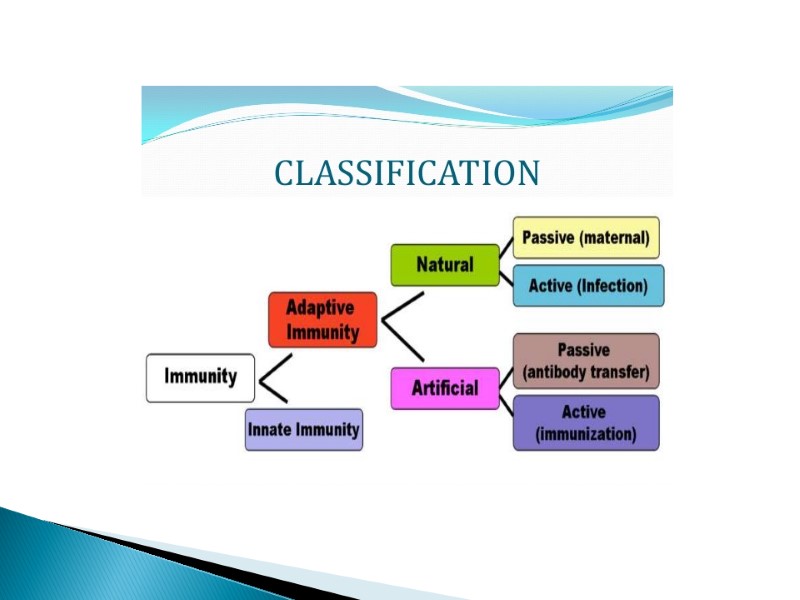
Classification of immune system Old and new classification







9472-classification_of_immune_system.ppt
- Количество слайдов: 26
 Classification of immune system Old and new classification
Classification of immune system Old and new classification






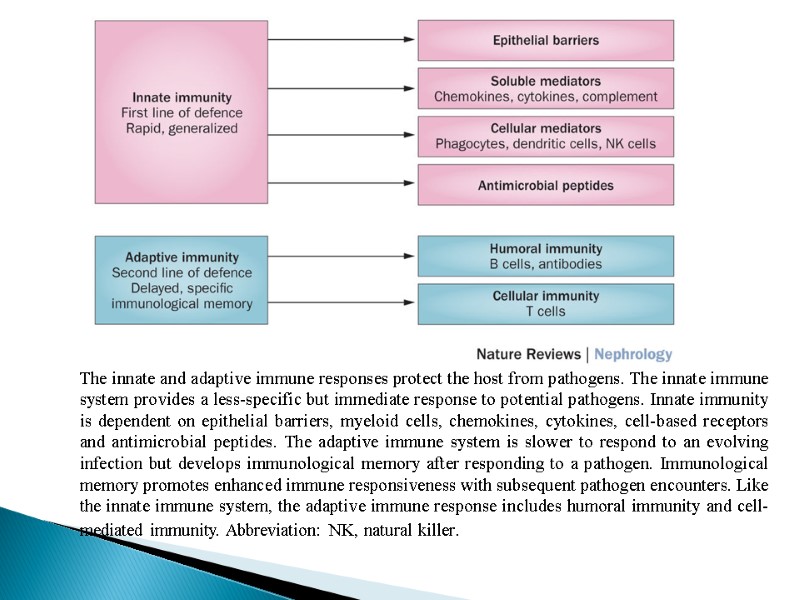
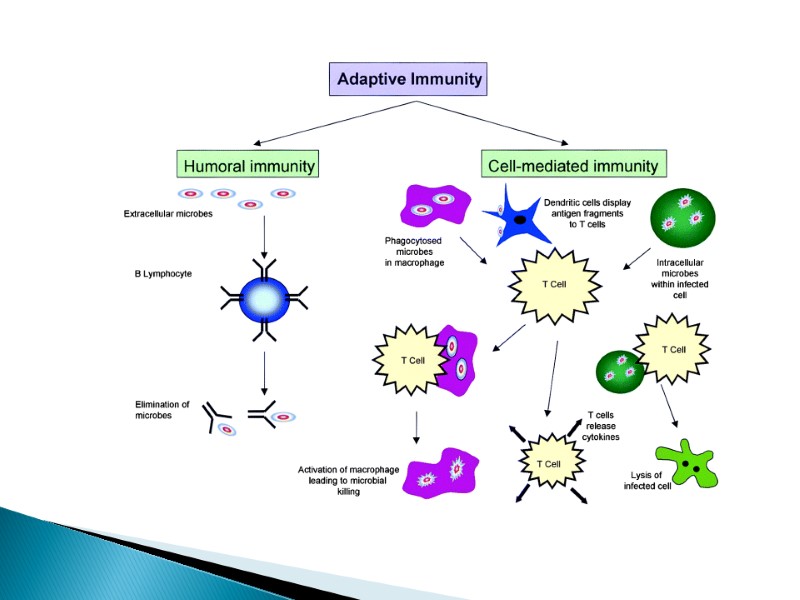
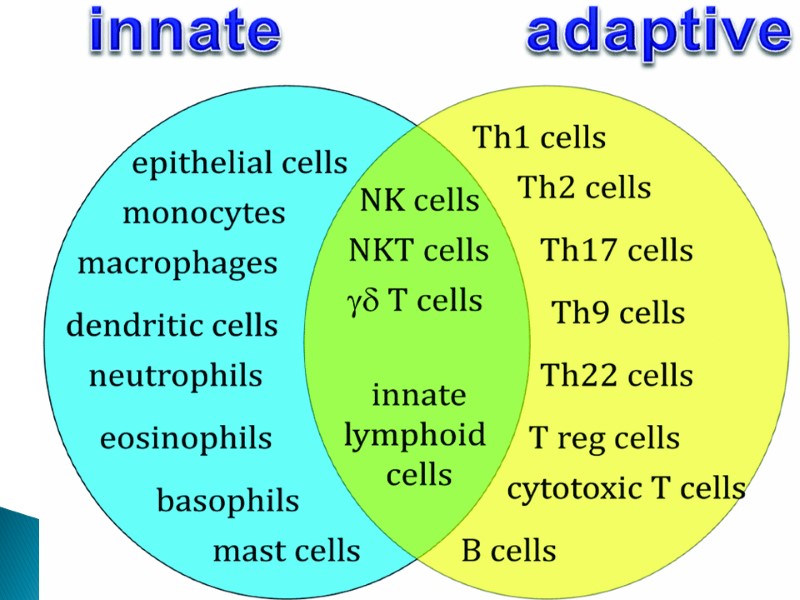
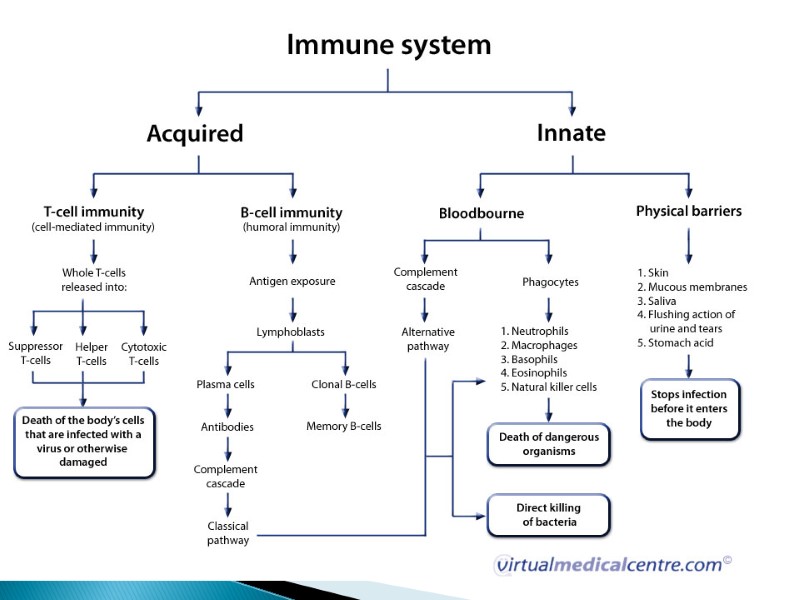
 The innate and adaptive immune responses protect the host from pathogens. The innate immune system provides a less-specific but immediate response to potential pathogens. Innate immunity is dependent on epithelial barriers, myeloid cells, chemokines, cytokines, cell-based receptors and antimicrobial peptides. The adaptive immune system is slower to respond to an evolving infection but develops immunological memory after responding to a pathogen. Immunological memory promotes enhanced immune responsiveness with subsequent pathogen encounters. Like the innate immune system, the adaptive immune response includes humoral immunity and cell-mediated immunity. Abbreviation: NK, natural killer.
The innate and adaptive immune responses protect the host from pathogens. The innate immune system provides a less-specific but immediate response to potential pathogens. Innate immunity is dependent on epithelial barriers, myeloid cells, chemokines, cytokines, cell-based receptors and antimicrobial peptides. The adaptive immune system is slower to respond to an evolving infection but develops immunological memory after responding to a pathogen. Immunological memory promotes enhanced immune responsiveness with subsequent pathogen encounters. Like the innate immune system, the adaptive immune response includes humoral immunity and cell-mediated immunity. Abbreviation: NK, natural killer.




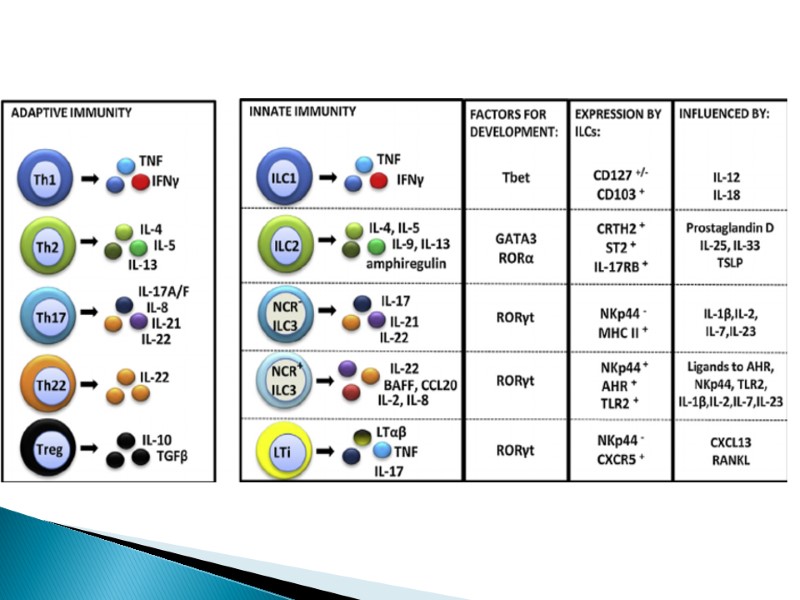
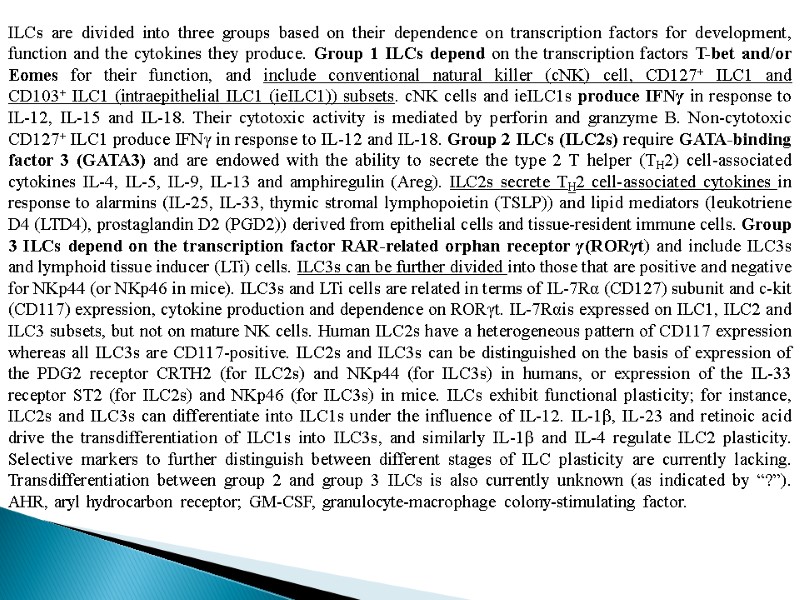
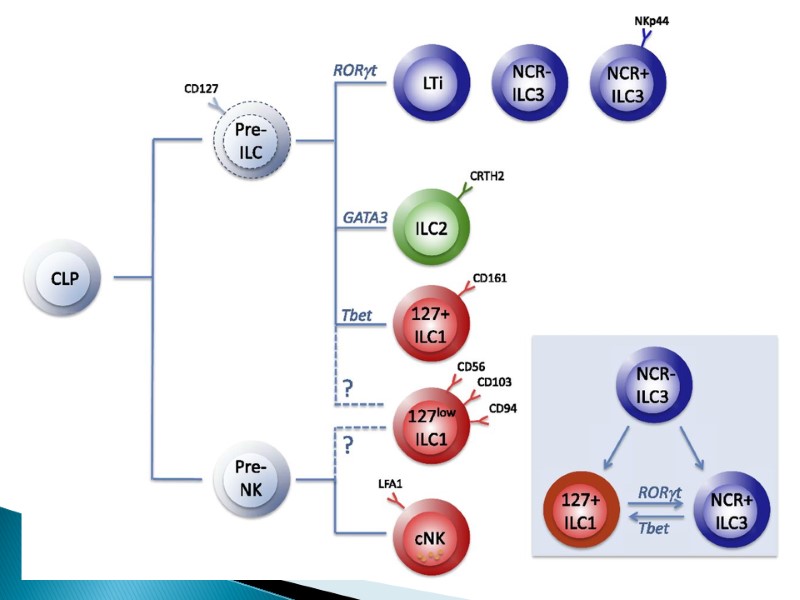
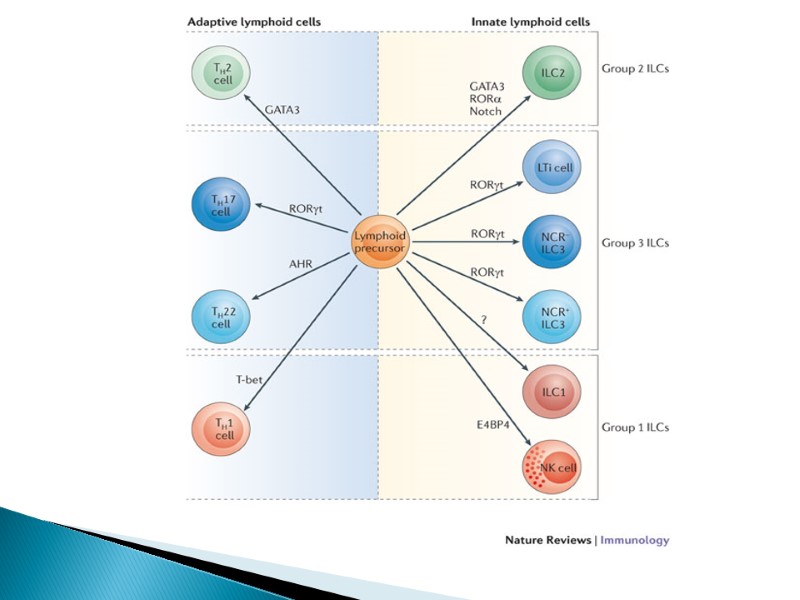
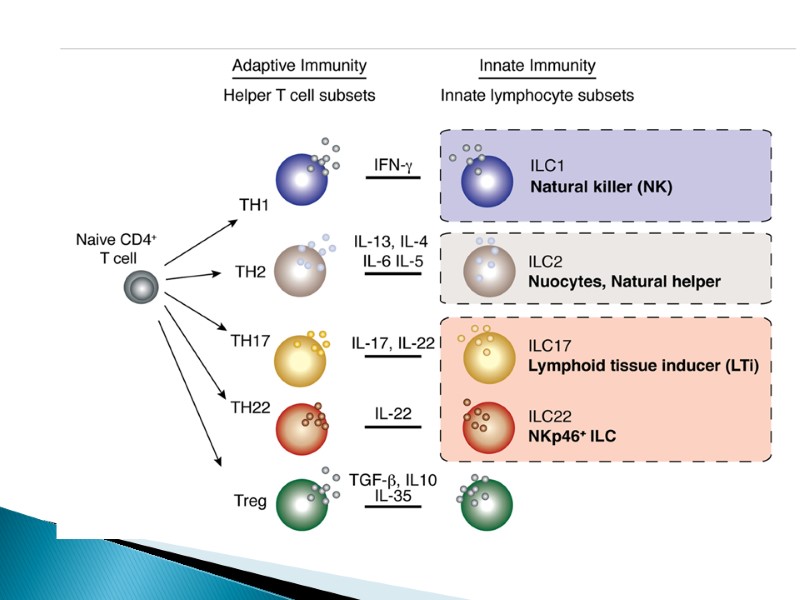
 ILCs are divided into three groups based on their dependence on transcription factors for development, function and the cytokines they produce. Group 1 ILCs depend on the transcription factors T-bet and/or Eomes for their function, and include conventional natural killer (cNK) cell, CD127+ ILC1 and CD103+ ILC1 (intraepithelial ILC1 (ieILC1)) subsets. cNK cells and ieILC1s produce IFNγ in response to IL-12, IL-15 and IL-18. Their cytotoxic activity is mediated by perforin and granzyme B. Non-cytotoxic CD127+ ILC1 produce IFNγ in response to IL-12 and IL-18. Group 2 ILCs (ILC2s) require GATA-binding factor 3 (GATA3) and are endowed with the ability to secrete the type 2 T helper (TH2) cell-associated cytokines IL-4, IL-5, IL-9, IL-13 and amphiregulin (Areg). ILC2s secrete TH2 cell-associated cytokines in response to alarmins (IL-25, IL-33, thymic stromal lymphopoietin (TSLP)) and lipid mediators (leukotriene D4 (LTD4), prostaglandin D2 (PGD2)) derived from epithelial cells and tissue-resident immune cells. Group 3 ILCs depend on the transcription factor RAR-related orphan receptor γ(RORγt) and include ILC3s and lymphoid tissue inducer (LTi) cells. ILC3s can be further divided into those that are positive and negative for NKp44 (or NKp46 in mice). ILC3s and LTi cells are related in terms of IL-7Rα (CD127) subunit and c-kit (CD117) expression, cytokine production and dependence on RORγt. IL-7Rαis expressed on ILC1, ILC2 and ILC3 subsets, but not on mature NK cells. Human ILC2s have a heterogeneous pattern of CD117 expression whereas all ILC3s are CD117-positive. ILC2s and ILC3s can be distinguished on the basis of expression of the PDG2 receptor CRTH2 (for ILC2s) and NKp44 (for ILC3s) in humans, or expression of the IL-33 receptor ST2 (for ILC2s) and NKp46 (for ILC3s) in mice. ILCs exhibit functional plasticity; for instance, ILC2s and ILC3s can differentiate into ILC1s under the influence of IL-12. IL-1β, IL-23 and retinoic acid drive the transdifferentiation of ILC1s into ILC3s, and similarly IL-1β and IL-4 regulate ILC2 plasticity. Selective markers to further distinguish between different stages of ILC plasticity are currently lacking. Transdifferentiation between group 2 and group 3 ILCs is also currently unknown (as indicated by “?”). AHR, aryl hydrocarbon receptor; GM-CSF, granulocyte-macrophage colony-stimulating factor.
ILCs are divided into three groups based on their dependence on transcription factors for development, function and the cytokines they produce. Group 1 ILCs depend on the transcription factors T-bet and/or Eomes for their function, and include conventional natural killer (cNK) cell, CD127+ ILC1 and CD103+ ILC1 (intraepithelial ILC1 (ieILC1)) subsets. cNK cells and ieILC1s produce IFNγ in response to IL-12, IL-15 and IL-18. Their cytotoxic activity is mediated by perforin and granzyme B. Non-cytotoxic CD127+ ILC1 produce IFNγ in response to IL-12 and IL-18. Group 2 ILCs (ILC2s) require GATA-binding factor 3 (GATA3) and are endowed with the ability to secrete the type 2 T helper (TH2) cell-associated cytokines IL-4, IL-5, IL-9, IL-13 and amphiregulin (Areg). ILC2s secrete TH2 cell-associated cytokines in response to alarmins (IL-25, IL-33, thymic stromal lymphopoietin (TSLP)) and lipid mediators (leukotriene D4 (LTD4), prostaglandin D2 (PGD2)) derived from epithelial cells and tissue-resident immune cells. Group 3 ILCs depend on the transcription factor RAR-related orphan receptor γ(RORγt) and include ILC3s and lymphoid tissue inducer (LTi) cells. ILC3s can be further divided into those that are positive and negative for NKp44 (or NKp46 in mice). ILC3s and LTi cells are related in terms of IL-7Rα (CD127) subunit and c-kit (CD117) expression, cytokine production and dependence on RORγt. IL-7Rαis expressed on ILC1, ILC2 and ILC3 subsets, but not on mature NK cells. Human ILC2s have a heterogeneous pattern of CD117 expression whereas all ILC3s are CD117-positive. ILC2s and ILC3s can be distinguished on the basis of expression of the PDG2 receptor CRTH2 (for ILC2s) and NKp44 (for ILC3s) in humans, or expression of the IL-33 receptor ST2 (for ILC2s) and NKp46 (for ILC3s) in mice. ILCs exhibit functional plasticity; for instance, ILC2s and ILC3s can differentiate into ILC1s under the influence of IL-12. IL-1β, IL-23 and retinoic acid drive the transdifferentiation of ILC1s into ILC3s, and similarly IL-1β and IL-4 regulate ILC2 plasticity. Selective markers to further distinguish between different stages of ILC plasticity are currently lacking. Transdifferentiation between group 2 and group 3 ILCs is also currently unknown (as indicated by “?”). AHR, aryl hydrocarbon receptor; GM-CSF, granulocyte-macrophage colony-stimulating factor.
 Transcription factors are one of the groups of proteins that read and interpret the genetic "blueprint" in the DNA. They bind to the DNA and help initiate a program of increased or decreased gene transcription. As such, they are vital for many important cellular processes. There are approximately 2600 proteins in the human genome that contain DNA-binding domains, and most of these are presumed to function as transcription factors, though other studies indicate it to be a smaller number. Therefore, approximately 10% of genes in the genome code for transcription factors, which makes this family the single largest family of human proteins. Furthermore, genes are often flanked by several binding sites for distinct transcription factors, and efficient expression of each of these genes requires the cooperative action of several different transcription factors. Hence, the combinatorial use of a subset of the approximately 2000 human transcription factors easily accounts for the unique regulation of each gene in the human genome during development.
Transcription factors are one of the groups of proteins that read and interpret the genetic "blueprint" in the DNA. They bind to the DNA and help initiate a program of increased or decreased gene transcription. As such, they are vital for many important cellular processes. There are approximately 2600 proteins in the human genome that contain DNA-binding domains, and most of these are presumed to function as transcription factors, though other studies indicate it to be a smaller number. Therefore, approximately 10% of genes in the genome code for transcription factors, which makes this family the single largest family of human proteins. Furthermore, genes are often flanked by several binding sites for distinct transcription factors, and efficient expression of each of these genes requires the cooperative action of several different transcription factors. Hence, the combinatorial use of a subset of the approximately 2000 human transcription factors easily accounts for the unique regulation of each gene in the human genome during development.








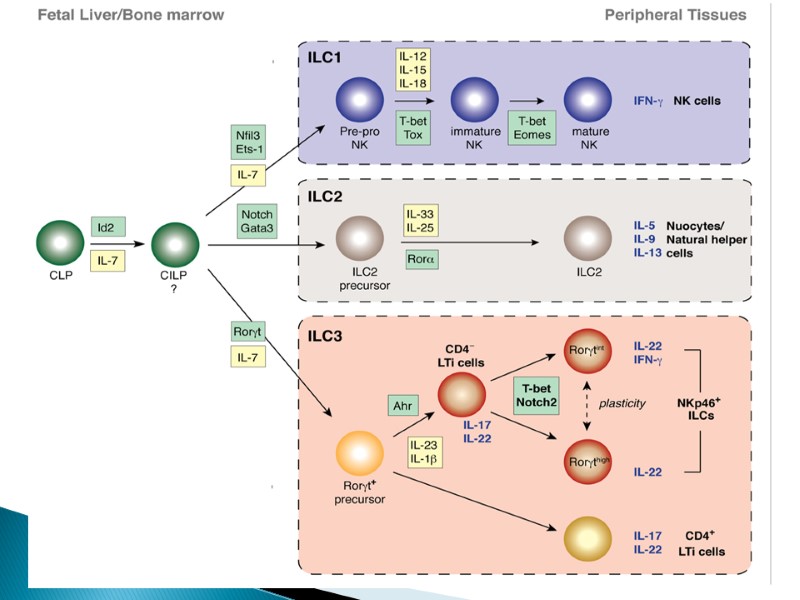
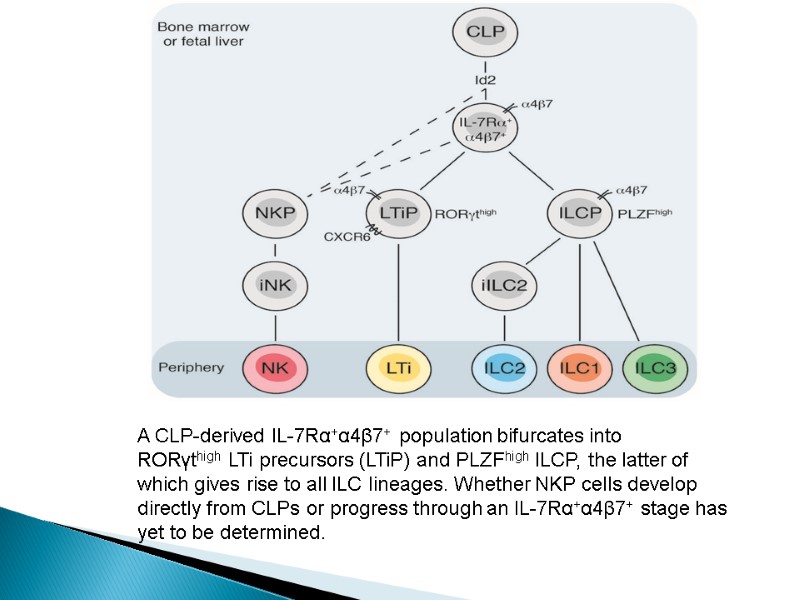
 A CLP-derived IL-7Rα+α4β7+ population bifurcates into RORγthigh LTi precursors (LTiP) and PLZFhigh ILCP, the latter of which gives rise to all ILC lineages. Whether NKP cells develop directly from CLPs or progress through an IL-7Rα+α4β7+ stage has yet to be determined.
A CLP-derived IL-7Rα+α4β7+ population bifurcates into RORγthigh LTi precursors (LTiP) and PLZFhigh ILCP, the latter of which gives rise to all ILC lineages. Whether NKP cells develop directly from CLPs or progress through an IL-7Rα+α4β7+ stage has yet to be determined.




